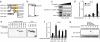Regulation of innate immune responses by DAI (DLM-1/ZBP1) and other DNA-sensing molecules - PubMed (original) (raw)
Regulation of innate immune responses by DAI (DLM-1/ZBP1) and other DNA-sensing molecules
Zhichao Wang et al. Proc Natl Acad Sci U S A. 2008.
Abstract
DNA, whether it is microbe-derived or host-derived, evokes immune responses when exposed to the cytosol of a cell. We previously reported that DNA-dependent activator of IFN regulatory factors (DAI), also referred to as DLM-1/ZBP1, functions as a DNA sensor that activates the innate immune system. In the present study, we examined the regulation of the complex DNA-sensing system by DAI and other molecules. We first show that DAI directly interacts with DNA in vitro and that it requires three DNA-binding domains for full activation in vivo. We also show that the artificially induced dimerization of DAI results in the DNA-independent activation of type I IFN genes, thereby providing a better understanding for the molecular basis of DAI activation. Furthermore, we provide evidence for the presence of additional DNA sensors, either positively or negatively regulating cytosolic DNA-mediated innate immune responses. These results in toto provide insights into the mechanism of DAI activation and reveal the complex regulatory mechanisms underlying DNA-mediated protective and pathologic immune responses.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
Recognition of DNAs by DAI protein. (a) Schematic illustration of DAI (DLM-1/ZBP1; DAI-WT) and its deletion mutants. Zα and Zβ DNA-binding domain (pink) and DNA-binding domain D3 (orange) are shown. DAI-ΔN2 is a deletion mutant of all three DNA-binding domains (14). (b) Coomassie brilliant blue (CBB) staining of GST or GST-DAI recombinant proteins. (c) Binding analysis of DAI protein with B-DNA. Recombinant DAI protein was incubated with biotin-conjugated B-DNA and with streptavidin (SA)-conjugated magnetic beads in the absence or presence of unconjugated B-DNA, poly(dG-dC)·poly(dC-dG), ISD, or poly(rI:rC) (0, 0.5, and 5.0 μg/ml; wedge above gels). DAI protein was analyzed by immunoblotting with anti-DAI antibody. (d) DNA length-dependent type I IFN production. L929 cells were transiently transfected with a plasmid vector for Renilla siRNA (Control) or for siRNA targeting DAI (siDAI#1). IFN-β mRNA induction was analyzed by qRT-PCR. Data are mean ± SD (n = 3). ND, not detected. *, P < 0.01 siDAI#1 versus control. (e) DNA pull-down analysis of DAI and DAI mutants. HEK293T cells were transiently transfected with plasmid for HA-tagged DAI-WT or deletion mutants as in a, and whole-cell lysate was incubated with biotin-conjugated B-DNA and SA-conjugated magnetic beads. (Left) Bound protein was analyzed by immunoblotting with anti-HA antibody. (Right) Input protein used for this assay is shown. (f) Type I IFN gene induction by DAI and DAI mutants. L929 cells that retrovirally expressed mock (Control), DAI-WT, or deletion mutants of DAI as in a were stimulated with 6 μg/ml B-DNA for 9 h. Induction of IFN-β mRNA was analyzed by qRT-PCR. Data are mean ± SD (n = 3). *, P < 0.01 DAI-WT or mutants versus control. (g) Interaction of DAI-WT and DAI mutants with IRF3. DAI-WT or deletion mutant of DAI as in a was transiently expressed in L929 cells with FLAG-tagged IRF3. Cells were stimulated with B-DNA for 2 h and analyzed by immunoprecipitation with anti-FLAG antibody, followed by immunoblotting with anti-HA (Upper) and anti-FLAG (Lower) antibodies.
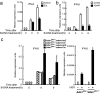
Fig. 2.
Dimerization/oligomerization of DAI activates immune response. (a) Dimer/oligomer formation of DAI by B-DNA stimulation. HA-tagged DAI (HA-DAI) and FLAG-tagged DAI (FLAG-DAI) were transiently coexpressed in L929 cells. The cells were stimulated with 6 μg/ml B-DNA for the indicated periods and analyzed by immunoprecipitation with anti-FLAG antibody, followed by immunoblotting with anti-HA (Upper) and anti-FLAG (Lower) antibodies. (b) Dimerization of DAI induces type I IFN production. HA-DAI or Fv (Fv-vector) or Fv-fused DAI (Fv-DAI) was transiently expressed in L929 cells. The cells were treated with AP20187 for the indicated periods, and the expressions of IFN-β (Top), IFN-α4 (Middle), and GAPDH (Bottom) mRNAs were assessed by RT-PCR. Results from B-DNA treatment on L929 cells are also shown. (c) DAI-WT, DAI-A1, or DAI-A2 was transiently expressed in L929 cells with either FLAG-TBK1 (Upper) or FLAG-IRF3 (Lower). These cells were stimulated with B-DNA and analyzed by immunoprecipitation with anti-FLAG antibody, followed by immunoblotting with anti-HA and anti-FLAG antibodies. The relative band intensities depicted in the graphs are of B-DNA-treated samples measured by a densitometer normalized to those of unstimulated sample.
Fig. 3.
Redundancy in the cytosolic DNA-sensing system. (a) L929 cells were transiently transfected with a plasmid vector for Renilla siRNA (Control) or for siRNA-targeting DAI (siDAI#1), stimulated with 6 μg/ml B-DNA for the indicated periods. (Left) Immunoblots with anti-DAI (Upper) and anti-β-actin (Lower) are shown. (Right) The induction of IFN-β mRNA in siRNA-expressing L929 cells after 6 h of treatment of B-DNA was assessed by qRT-PCR. Data are mean ± SD (n = 3). *, P < 0.01 siDAI#1 versus control. (b) MEFs were transiently transfected with Renilla siRNA (Control) or siDAI#1 plasmid vector, stimulated with B-DNA. (Left) Immunoblots with anti-DAI (Upper) and anti-β-actin (Lower) are shown. (Center and Right) The induction of IFN-β mRNA in siRNA-expressing MEFs after B-DNA stimulation (Center) or 10 μg/ml of ISD stimulation (Right) was assessed by qRT-PCR. Data are mean ± SD. (n = 3). *, P < 0.001 siDAI#1 versus control.
Fig. 4.
Negative regulation of DNA-sensing system. (a and b) MEFs that retrovirally expressed mock (Control, white bars), E3L (a) (black bars), or ADAR1 (b) (black bars) were stimulated with 6 μg/ml B-DNA for the indicated periods. Induction of IFN-β mRNA was analyzed by qRT-PCR. *, P < 0.01; E3L versus control (a) or ADAR1 versus control (b). Data are mean ± SD (n = 3). (c) Adar1 flox/+ and Adar1 flox/− MEFs were stimulated with B-DNA (Left) for the indicated periods or infected with 1 moi of HSV-1 (Right) for 18 h. Induction of IFN-β mRNA was analyzed by qRT-PCR. *, P < 0.01 Adar1 flox/−-adeno-Cre versus Adar1 flox/+-adeno-Cre. Data are mean ± SD (n = 3).
Similar articles
- DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response.
Takaoka A, Wang Z, Choi MK, Yanai H, Negishi H, Ban T, Lu Y, Miyagishi M, Kodama T, Honda K, Ohba Y, Taniguchi T. Takaoka A, et al. Nature. 2007 Jul 26;448(7152):501-5. doi: 10.1038/nature06013. Epub 2007 Jul 8. Nature. 2007. PMID: 17618271 - Cytosolic DNA recognition for triggering innate immune responses.
Takaoka A, Taniguchi T. Takaoka A, et al. Adv Drug Deliv Rev. 2008 Apr 29;60(7):847-57. doi: 10.1016/j.addr.2007.12.002. Epub 2007 Dec 31. Adv Drug Deliv Rev. 2008. PMID: 18280611 Review. - DNA-dependent activator of interferon-regulatory factors (DAI) promotes lupus nephritis by activating the calcium pathway.
Zhang W, Zhou Q, Xu W, Cai Y, Yin Z, Gao X, Xiong S. Zhang W, et al. J Biol Chem. 2013 May 10;288(19):13534-50. doi: 10.1074/jbc.M113.457218. Epub 2013 Apr 3. J Biol Chem. 2013. PMID: 23553627 Free PMC article. - DNA sensors in innate immune system.
Takaoka A, Shinohara S. Takaoka A, et al. Uirusu. 2008 Jun;58(1):37-46. doi: 10.2222/jsv.58.37. Uirusu. 2008. PMID: 19122387 Review. - Regulation of the cytosolic DNA-sensing system in innate immunity: a current view.
Yanai H, Savitsky D, Tamura T, Taniguchi T. Yanai H, et al. Curr Opin Immunol. 2009 Feb;21(1):17-22. doi: 10.1016/j.coi.2009.01.005. Curr Opin Immunol. 2009. PMID: 19362700 Review.
Cited by
- Innate immune detection of microbial nucleic acids.
Gürtler C, Bowie AG. Gürtler C, et al. Trends Microbiol. 2013 Aug;21(8):413-20. doi: 10.1016/j.tim.2013.04.004. Epub 2013 May 29. Trends Microbiol. 2013. PMID: 23726320 Free PMC article. Review. - Extrachromosomal histone H2B mediates innate antiviral immune responses induced by intracellular double-stranded DNA.
Kobiyama K, Takeshita F, Jounai N, Sakaue-Sawano A, Miyawaki A, Ishii KJ, Kawai T, Sasaki S, Hirano H, Ishii N, Okuda K, Suzuki K. Kobiyama K, et al. J Virol. 2010 Jan;84(2):822-32. doi: 10.1128/JVI.01339-09. Epub 2009 Nov 11. J Virol. 2010. PMID: 19906922 Free PMC article. - The porcine innate immune system: an update.
Mair KH, Sedlak C, Käser T, Pasternak A, Levast B, Gerner W, Saalmüller A, Summerfield A, Gerdts V, Wilson HL, Meurens F. Mair KH, et al. Dev Comp Immunol. 2014 Aug;45(2):321-43. doi: 10.1016/j.dci.2014.03.022. Epub 2014 Apr 4. Dev Comp Immunol. 2014. PMID: 24709051 Free PMC article. Review. - Solution structure of the Zbeta domain of human DNA-dependent activator of IFN-regulatory factors and its binding modes to B- and Z-DNAs.
Kim K, Khayrutdinov BI, Lee CK, Cheong HK, Kang SW, Park H, Lee S, Kim YG, Jee J, Rich A, Kim KK, Jeon YH. Kim K, et al. Proc Natl Acad Sci U S A. 2011 Apr 26;108(17):6921-6. doi: 10.1073/pnas.1014898107. Epub 2011 Apr 6. Proc Natl Acad Sci U S A. 2011. PMID: 21471454 Free PMC article. - Protect this house: cytosolic sensing of viruses.
McFadden MJ, Gokhale NS, Horner SM. McFadden MJ, et al. Curr Opin Virol. 2017 Feb;22:36-43. doi: 10.1016/j.coviro.2016.11.012. Epub 2016 Dec 9. Curr Opin Virol. 2017. PMID: 27951430 Free PMC article. Review.
References
- Tokunaga T, Yamamoto T, Yamamoto S. How BCG led to the discovery of immunostimulatory DNA. Jpn J Infect Dis. 1999;52:1–11. - PubMed
- Ishii KJ, et al. A Toll-like receptor-independent antiviral response induced by double-stranded B-form DNA. Nat Immunol. 2006;7:40–48. - PubMed
- Stetson DB, Medzhitov R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 2006;24:93–103. - PubMed
- Yoshida H, Okabe Y, Kawane K, Fukuyama H, Nagata S. Lethal anemia caused by interferon-β produced in mouse embryos carrying undigested DNA. Nat Immunol. 2005;6:49–56. - PubMed
- Napirei M, et al. Features of systemic lupus erythematosus in Dnase1-deficient mice. Nat Genet. 2000;25:177–181. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases