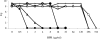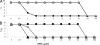In vitro selection and characterization of ceftobiprole-resistant methicillin-resistant Staphylococcus aureus - PubMed (original) (raw)
In vitro selection and characterization of ceftobiprole-resistant methicillin-resistant Staphylococcus aureus
Ritu Banerjee et al. Antimicrob Agents Chemother. 2008 Jun.
Abstract
Methicillin-resistant Staphylococcus aureus (MRSA) is resistant to beta-lactam antibiotics because it expresses penicillin-binding protein 2a (PBP2a), a low-affinity penicillin-binding protein. An investigational broad-spectrum cephalosporin, ceftobiprole (BPR), binds PBP2a with high affinity and is active against MRSA. We hypothesized that BPR resistance could be mediated by mutations in mecA, the gene encoding PBP2a. We selected BPR-resistant mutants by passage in high-volume broth cultures containing subinhibitory concentrations of BPR. We used strain COLnex (which lacks chromosomal mecA) transformed with pAW8 (a plasmid vector only), pYK20 (a plasmid carrying wild-type mecA), or pYK21 (a plasmid carrying a mutant mecA gene corresponding to five PBP2a mutations). All strains became resistant to BPR by day 9 of passaging, but MICs continued to increase until day 21. MICs increased 256-fold (from 1 to 256 microg/ml) for pAW8, 32-fold (from 4 to 128 microg/ml) for pYK20, and 8-fold (from 16 to 128 mug/ml) for pYK21. Strains carrying wild-type or mutant mecA developed six (pYK20 transformants) or four (pYK21 transformants) new mutations in mecA. The transformation of COLnex with a mecA mutant plasmid conferred BPR resistance, and the loss of mecA converted resistant strains into susceptible ones. Modeling studies predicted that several of the mecA mutations altered BPR binding; other mutations may have mediated resistance by influencing interactions with other proteins. Multiple mecA mutations were associated with BPR resistance in MRSA. BPR resistance also developed in the strain lacking mecA, suggesting a role for chromosomal genes.
Figures
FIG. 1.
BPR resistance developed in multiple steps during the serial passage of strains COLnex(pAW8) (filled squares), COLnex(pYK20) (filled triangles), and COLnex(pYK21) (open circles). The highest BPR concentration in which strains grew each day is shown on the y axis. The inset is a close-up view of results from the first 2 weeks of passaging. The dashed line represents the preliminary BPR breakpoint of 4 μg/ml.
FIG. 2.
Population analyses showing BPR susceptibilities of prepassage strains [filled squares, COLnex(pAW8); filled triangles, COLnex(pYK20); and filled circles, COLnex(pYK21)] and strains passaged in BPR for 28 days [open squares, COLnex(pAW8)D28; open triangles, COLnex(pYK20)D28; and open circles, COLnex(pYK21)D28]. The y axis indicates the number of cells (expressed as the log10 number of CFU per milliliter) growing on BPR-containing agar.
FIG. 3.
Growth curves of strains before and after BPR passage. Filled squares, COLnex(pAW8); open squares, COLnex(pAW8)D28; filled triangles, COLnex(pYK20); open triangles, COLnex(pYK20)D28; filled circles, COLnex(pYK21); and open circles COLnex(pYK21)D28. OD578, optical density at 578 nm.
FIG. 4.
Schematic of PBP2a and amino acid substitutions in BPR-passaged strains containing plasmid-carried mecA, COLnex(pYK20) and COLnex(pYK21). The day of serial passage and the corresponding amino acid changes identified are shown. In the schematic, vertical black lines indicate three penicillin-binding motifs, the arrowhead denotes a transmembrane anchor, the speckled region denotes the non-penicillin-binding domain (nonPBD), and the diagonally striped region denotes the transpeptidase domain. Underlined amino acid substitutions arose independently in derivatives of both COLnex(pYK20) and COLnex(pYK21). D0, D13, D15, and D28, days 0, 13, 15, and 28.
FIG. 5.
(A) Population analyses of COLnex transformed with plasmids derived from BPR-passaged strains: COLnex(pAW8)T (filled squares), COLnex(pYK20)T (filled triangles), and COLnex(pYK21)T (open circles). The y axis indicates the number of cells (expressed as the log10 number of CFU per milliliter) growing on BPR-containing agar. (B) Population analyses of BPR-resistant strains cured of plasmid by passage in the absence of tetracycline: COLnex(pAW8)c (filled squares), COLnex(pYK20)c (filled triangles), and COLnex(pYK21)c (open circles). The y axis indicates the number of cells (expressed as the log10 number of CFU per milliliter) growing on BPR-containing agar.
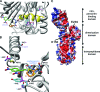
FIG. 6.
Structural perspectives of BPR-resistant PBP2a mutant forms. (A and B) Molecular modeling of mutant forms of PBP2a bound to the cephalosporin nitrocefin showing nitrocefin in blue (cephalosporin core structure), orange (variable group R1), and green (group R2); the native protein backbone and residues in gray; residues subject to mutation in cyan (native residue) and purple (corresponding mutant residue); and noncarbon atoms in CPK. (A) Group 1 mutations (F467Y, V470E, and I563T) likely affect BPR binding by perturbing helix α2 (shown as a yellow ribbon) by introducing adjacent polar or charged groups into the hydrophobic interior of PBP2a. (B) Group 2 substitutions probably lower BPR-binding affinity by directly affecting the PBP2a active site. These substitutions include Y446L, predicted to disrupt van der Waals contacts and pi bond stacking (π-π) interactions between PBP2a and conjugated double-bond systems of cephalosporin R2 groups; E447K, which may interact electrostatically with E460 to reposition Y446; S649A, predicted to destabilize the alpha helix bearing M641 that interacts with aromatic or hydrophobic R2 groups; and S643N, which may alter the polarity at the active-site entrance. (C) APBS modeling of PBP2a surface charge potential shows native residues E150 and E239 located within an extended swath of negative charge in the putative dimerization domain (the APBS surface is colored blue, white, and red, corresponding to values of +15, 0, and −15 kT/e, respectively, with shading by linear interpolation). Each of these residues is mutated, respectively, in pYK20 and pYK21, possibly contributing to BPR resistance by influencing protein-protein interactions.
Similar articles
- Whole-Genome Sequencing of Methicillin-Resistant Staphylococcus aureus Resistant to Fifth-Generation Cephalosporins Reveals Potential Non-mecA Mechanisms of Resistance.
Greninger AL, Chatterjee SS, Chan LC, Hamilton SM, Chambers HF, Chiu CY. Greninger AL, et al. PLoS One. 2016 Feb 18;11(2):e0149541. doi: 10.1371/journal.pone.0149541. eCollection 2016. PLoS One. 2016. PMID: 26890675 Free PMC article. - Comparative study of the susceptibilities of major epidemic clones of methicillin-resistant Staphylococcus aureus to oxacillin and to the new broad-spectrum cephalosporin ceftobiprole.
Chung M, Antignac A, Kim C, Tomasz A. Chung M, et al. Antimicrob Agents Chemother. 2008 Aug;52(8):2709-17. doi: 10.1128/AAC.00266-08. Epub 2008 May 27. Antimicrob Agents Chemother. 2008. PMID: 18505853 Free PMC article. - The mecA homolog mecC confers resistance against β-lactams in Staphylococcus aureus irrespective of the genetic strain background.
Ballhausen B, Kriegeskorte A, Schleimer N, Peters G, Becker K. Ballhausen B, et al. Antimicrob Agents Chemother. 2014 Jul;58(7):3791-8. doi: 10.1128/AAC.02731-13. Epub 2014 Apr 21. Antimicrob Agents Chemother. 2014. PMID: 24752255 Free PMC article. - A Review on Five and Six-Membered Heterocyclic Compounds Targeting the Penicillin-Binding Protein 2 (PBP2A) of Methicillin-Resistant Staphylococcus aureus (MRSA).
Ambade SS, Gupta VK, Bhole RP, Khedekar PB, Chikhale RV. Ambade SS, et al. Molecules. 2023 Oct 10;28(20):7008. doi: 10.3390/molecules28207008. Molecules. 2023. PMID: 37894491 Free PMC article. Review. - In-vitro profile of a new beta-lactam, ceftobiprole, with activity against methicillin-resistant Staphylococcus aureus.
Jones ME. Jones ME. Clin Microbiol Infect. 2007 Jun;13 Suppl 2:17-24. doi: 10.1111/j.1469-0691.2007.01722.x. Clin Microbiol Infect. 2007. PMID: 17488372 Review.
Cited by
- Missense mutations in PBP2A Affecting ceftaroline susceptibility detected in epidemic hospital-acquired methicillin-resistant Staphylococcus aureus clonotypes ST228 and ST247 in Western Switzerland archived since 1998.
Kelley WL, Jousselin A, Barras C, Lelong E, Renzoni A. Kelley WL, et al. Antimicrob Agents Chemother. 2015 Apr;59(4):1922-30. doi: 10.1128/AAC.04068-14. Epub 2015 Jan 12. Antimicrob Agents Chemother. 2015. PMID: 25583724 Free PMC article. - Identification of Staphylococcus aureus Penicillin Binding Protein 4 (PBP4) Inhibitors.
Young M, Walsh DJ, Masters E, Gondil VS, Laskey E, Klaczko M, Awad H, McGrath J, Schwarz EM, Melander C, Dunman PM. Young M, et al. Antibiotics (Basel). 2022 Oct 4;11(10):1351. doi: 10.3390/antibiotics11101351. Antibiotics (Basel). 2022. PMID: 36290009 Free PMC article. - An experiment-informed signal transduction model for the role of the Staphylococcus aureus MecR1 protein in β-lactam resistance.
Belluzo BS, Abriata LA, Giannini E, Mihovilcevic D, Dal Peraro M, Llarrull LI. Belluzo BS, et al. Sci Rep. 2019 Dec 20;9(1):19558. doi: 10.1038/s41598-019-55923-z. Sci Rep. 2019. PMID: 31862951 Free PMC article. - Molecular basis and phenotype of methicillin resistance in Staphylococcus aureus and insights into new beta-lactams that meet the challenge.
Llarrull LI, Fisher JF, Mobashery S. Llarrull LI, et al. Antimicrob Agents Chemother. 2009 Oct;53(10):4051-63. doi: 10.1128/AAC.00084-09. Epub 2009 May 26. Antimicrob Agents Chemother. 2009. PMID: 19470504 Free PMC article. Review. No abstract available. - In Vitro Antibacterial Activity of Ceftobiprole and Comparator Compounds against Nation-Wide Bloodstream Isolates and Different Sequence Types of MRSA.
Li L, Zhou W, Chen Y, Shen P, Xiao Y. Li L, et al. Antibiotics (Basel). 2024 Feb 7;13(2):165. doi: 10.3390/antibiotics13020165. Antibiotics (Basel). 2024. PMID: 38391551 Free PMC article.
References
- Appelbaum, P. C. 2006. MRSA—the tip of the iceberg. Clin. Microbiol. Infect. 12(Suppl. 2):3-10. - PubMed
- Cornell, D. W., P. Cieplak, and C. I. Bayly. 1995. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc. 117:5179.
- Davies, T. A., M. G. P. Page, W. Shang, T. Andrew, M. Kania, and K. Bush. 2007. Binding of ceftobiprole and comparators to the penicillin-binding proteins of Escherichia coli, Pseudomonas aeruginosa, Staphylococcus aureus, and Streptococcus pneumoniae. Antimicrob. Agents Chemother. 51:2621-2624. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical