Evidence that strong positive selection drives neofunctionalization in the tandemly duplicated polyhomeotic genes in Drosophila - PubMed (original) (raw)
Evidence that strong positive selection drives neofunctionalization in the tandemly duplicated polyhomeotic genes in Drosophila
Steffen Beisswanger et al. Proc Natl Acad Sci U S A. 2008.
Abstract
The polyhomeotic (ph) locus in Drosophila melanogaster consists of the two tandemly duplicated genes ph-d (distal) and ph-p (proximal). They code for transcriptional repressors belonging to the Polycomb group proteins, which regulate homeotic genes and hundreds of other loci. Although the duplication of ph occurred at least 25 million to 30 million years ago, both copies are very similar to each other at both the DNA and the protein levels, probably because of the action of frequent gene conversion. Despite this homogenizing force, differential regulation of both transcriptional units suggests that the functions of the duplicates have begun to diverge. Here, we provide evidence that this functional divergence is driven by positive selection. Based on resequencing of an approximately 30-kb region around the ph locus in an African sample of D. melanogaster X chromosomes, we identified a selective sweep, estimated its age and the strength of selection, and mapped the target of selection to a narrow interval of the ph-p gene. This noncoding region contains a large intron with several regulatory elements that are absent in the ph-d duplicate. Our results suggest that neofunctionalization has been achieved in the Drosophila ph genes through the action of strong positive selection and the inactivation of gene conversion in part of the gene.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
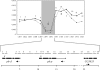
Fig. 1.
Illustration of the _ph-d_–CG3835 region. (Upper) Modified from ref. , the entire wapl region is shown. The wapl fragment is indicated by a star. It harbors no variation in the European sample and was detected in a previous genome scan (34). The numbers on the x axis are the absolute genomic position (in Mb), and those on the y axis are nucleotide variability in the African sample (the solid line corresponds to θ and the dashed line corresponds to π). The arrow indicates the position of the target of selection predicted by Beisswanger et al. (17). (Lower) The region investigated in detail in this study. Exons are illustrated by thick black lines and introns by thin black lines. The direction of transcription is indicated by dotted arrows below each gene. Sequenced fragments are denoted by gray lines and numbers. Gray arrow heads indicate the position of the three SNPs shared between both ph copies. These SNPs are located within a region of 135 bp.
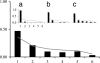
Fig. 2.
Site frequency spectra. The folded frequency spectrum of 27 loci randomly chosen from ref. . (Inset) Shown are the spectra of the ph-p intron and the 5′ UTR (a), the first half of the _ph-p_–CG3835 intergenic region (b), and the second half of that region (c). The gray lines indicate the equilibrium frequencies.
Similar articles
- The polyhomeotic locus of Drosophila melanogaster is transcriptionally and post-transcriptionally regulated during embryogenesis.
Hodgson JW, Cheng NN, Sinclair DA, Kyba M, Randsholt NB, Brock HW. Hodgson JW, et al. Mech Dev. 1997 Aug;66(1-2):69-81. doi: 10.1016/s0925-4773(97)00091-9. Mech Dev. 1997. PMID: 9376325 - P-Element insertion at the polyhomeotic gene leads to formation of a novel chimeric protein that negatively regulates yellow gene expression in P-element-induced alleles of Drosophila melanogaster.
Belenkaya T, Soldatov A, Nabirochkina E, Biryukova I, Georgieva S, Georgiev P. Belenkaya T, et al. Genetics. 1998 Oct;150(2):687-97. doi: 10.1093/genetics/150.2.687. Genetics. 1998. PMID: 9755200 Free PMC article. - Transcriptional silencing of homeotic genes in Drosophila.
Bienz M, Müller J. Bienz M, et al. Bioessays. 1995 Sep;17(9):775-84. doi: 10.1002/bies.950170907. Bioessays. 1995. PMID: 8763830 Review. - From genetics to epigenetics: the tale of Polycomb group and trithorax group genes.
Grimaud C, Nègre N, Cavalli G. Grimaud C, et al. Chromosome Res. 2006;14(4):363-75. doi: 10.1007/s10577-006-1069-y. Chromosome Res. 2006. PMID: 16821133 Review.
Cited by
- Patterns of nucleotide diversity at the regions encompassing the Drosophila insulin-like peptide (dilp) genes: demography vs. positive selection in Drosophila melanogaster.
Guirao-Rico S, Aguadé M. Guirao-Rico S, et al. PLoS One. 2013;8(1):e53593. doi: 10.1371/journal.pone.0053593. Epub 2013 Jan 7. PLoS One. 2013. PMID: 23308258 Free PMC article. - Molecular evolution of teleost neural isozymes.
Auld RR, Quattro JM, Merritt TJ. Auld RR, et al. J Mol Evol. 2012 Dec;75(5-6):198-213. doi: 10.1007/s00239-012-9532-1. Epub 2012 Nov 25. J Mol Evol. 2012. PMID: 23183893 - Genome-Wide Dissection of the Heat Shock Transcription Factor Family Genes in Arachis.
Wang P, Song H, Li C, Li P, Li A, Guan H, Hou L, Wang X. Wang P, et al. Front Plant Sci. 2017 Feb 6;8:106. doi: 10.3389/fpls.2017.00106. eCollection 2017. Front Plant Sci. 2017. PMID: 28220134 Free PMC article. - Accelerated evolution and coevolution drove the evolutionary history of AGPase sub-units during angiosperm radiation.
Corbi J, Dutheil JY, Damerval C, Tenaillon MI, Manicacci D. Corbi J, et al. Ann Bot. 2012 Mar;109(4):693-708. doi: 10.1093/aob/mcr303. Epub 2012 Feb 2. Ann Bot. 2012. PMID: 22307567 Free PMC article. - Evolution of Type II Antifreeze Protein Genes in Teleost Fish: A Complex Scenario Involving Lateral Gene Transfers and Episodic Directional Selection.
Sorhannus U. Sorhannus U. Evol Bioinform Online. 2012;8:535-44. doi: 10.4137/EBO.S9976. Epub 2012 Sep 17. Evol Bioinform Online. 2012. PMID: 23032610 Free PMC article.
References
- Ohno S. Evolution by Gene Duplication. Berlin: Springer; 1970.
- Ohta T. Evolution and Variation of Multigene Families: Lecture Notes in Biomathematics 37. Berlin: Springer; 1980.
- Walsh JB. Population-genetic models of the fates of duplicate genes. Genetica. 2003;118:279–294. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases