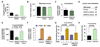TGFbeta primes breast tumors for lung metastasis seeding through angiopoietin-like 4 - PubMed (original) (raw)
TGFbeta primes breast tumors for lung metastasis seeding through angiopoietin-like 4
David Padua et al. Cell. 2008.
Abstract
Cells released from primary tumors seed metastases to specific organs by a nonrandom process, implying the involvement of biologically selective mechanisms. Based on clinical, functional, and molecular evidence, we show that the cytokine TGFbeta in the breast tumor microenvironment primes cancer cells for metastasis to the lungs. Central to this process is the induction of angiopoietin-like 4 (ANGPTL4) by TGFbeta via the Smad signaling pathway. TGFbeta induction of Angptl4 in cancer cells that are about to enter the circulation enhances their subsequent retention in the lungs, but not in the bone. Tumor cell-derived Angptl4 disrupts vascular endothelial cell-cell junctions, increases the permeability of lung capillaries, and facilitates the trans-endothelial passage of tumor cells. These results suggest a mechanism for metastasis whereby a cytokine in the primary tumor microenvironment induces the expression of another cytokine in departing tumor cells, empowering these cells to disrupt lung capillary walls and seed pulmonary metastases.
Figures
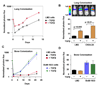
Figure 1. The TBRS associates with breast cancer metastasis in humans
(A) The indicated epithelial cell lines were incubated for 3 h with TGFβ and then total RNA was subjected to microarray analysis. The heat map represents the change in expression levels of the 153 genes within the TBRS. (B) TBRS status was assessed in a MSK/EMC cohort of 368 primary breast cancer tumors with known lung or bone metastatic outcomes. Red denotes a strong correlation between individual tumor gene expression profiles and the TBRS while blue indicates no correlation. Estrogen receptor α(ER) expression status is also indicated. Blue and red marks above the heat map indicate tumors that at any time developed bone or lung metastases, respectively. (C) Kaplan-Meier curves representing the probability of cumulative lung (left panel) or bone (right panel) metastasis-free survival for this cohort. Tumors are categorized according to their TBRS and ER status. The P values for the ER-negative tumor comparisons are shown. (D) Hierarchical clustering was performed on the MSK/EMC cohort with the indicated pathological and genomic markers including the TBRS, the lung metastasis signature (LMS), the wound response signature (Wound), the 70-gene prognosis signature (70-gene), size (Size >2cm), the basal molecular subtype (Basal), and the ER status. Red marks above the map indicate tumors that developed lung metastasis. (E) Lung metastasis-free survival restricted to patients with ER-negative tumors. Patients were categorized according to their TBRS and LMS status. P value shown for the LMS+ tumor comparisons.
Figure 2. TGFβ signaling enhances mammary tumor dissemination to the lungs
(A) Schematic of lung metastasis assay from an orthotopic breast cancer inoculation. (B) Immunoblots using indicated antibodies were performed on whole-cell extracts from control, Smad4 knockdown, and Smad4-Rescue LM2 cells. (C) Mice injected with 5×105 cells into the fourth mammary fat pads were measured for tumor size at day 28. n=14; error bars indicated s.e.m (D) Blood from tumor-bearing mice was isolated and red blood cells lysed. RNA from the remaining cells was extracted for qRT-PCR. The presence of circulating tumor cells was assessed as a function of human-specific GAPDH expression relative to murine β2-microglobulin, in 3 mL of mouse blood perfusate. n=8; error bars indicate s.e.m. (E) Bioluminescent quantification of lung seeding elicited from orthotopically implanted breast tumors. Orthotopic tumors were grown to approximately 300mm3, mastectomies were performed and lung seeding was quantified using bioluminescence imaging seven days later. n=7–15; error bars indicated s.e.m; p-values calculated using the one-tailed unpaired t-test. (F) Representative bioluminescent images (inset) and lung histology of mice with the median value of bioluminescence in each experimental group in (D). Breast cancer cells were stained with human vimentin.
Figure 3. TGFβ primes tumor cells for metastatic seeding of the lungs
(A) LM2 cells and a clinically-derived pleural effusion sample (CN34.2A) were pretreated with TGFβ for 6 h. LM2 (2×105) and CN34.2A (4×105) cells were injected into the lateral tail vein and lung colonization was analyzed by in vivo bioluminescence imaging. (B) Bar graph represents 24h time point measurements of the normalized photon flux from animals injected with either LM2 or CN34.2A cells. In vivo bioluminescent mouse images shown are from representative animals. n=7 for the LM2 experiment while n=6 for the CN34.2A experiment; error bars indicate s.e.m; p-values calculated using the one-tailed unpaired t-test. (C) Bone colonization assays were performed by intracardiac injection of LM2 or BoM-1833 cells (3×104). Samples were pretreated with 100 pM of TGFβ for 6 h and compared to an untreated control. Plot represents normalized photon flux from mouse hindlimbs (D) Bar graphs represent seven-day time point analysis of the normalized photon flux from the mouse hind limbs. n=8; error bars indicate s.e.m.
Figure 4. The TBRS/LMS gene ANGPTL4 is a Smad-dependent TGFβ target
(A) Microarray and qRT-PCR analysis for the four epithelial cell lines treated with TGFβ. Fold change values for the TGFβ induction of ANGPTL4 are indicated. (B) Box-and-whisker plot comparing ANGPTL4 and NEDD9 TBRS-negative and -positive ER-negative tumors from the MSK/EMC cohorts. P value was calculated using the Wilcoxon rank sum test. (C) TGFβ-induced changes in the mRNA expression of LMS genes in a panel of clinically derived pleural effusion samples and LM2 cells. Cells were treated with 100 pM of TGFβ for 3 h and analyzed by qRT-PCR using primers for the indicated genes. ER status for each breast cancer patient is designated. (D) LM2 breast cancer cells were treated with 100pM of TGFβ for the indicated lengths of time and ANGPTL4 mRNA levels were analyzed using qRT-PCR. (E) Treatment of LM2 (left panel) and pleural effusion-derived CN37 sample (right panel) with TGFβ and the TGFβ-receptor kinase inhibitor, SB431542. qRT-PCR expression levels are shown relative to the untreated control sample. (F) MDA-231, LM2 control, LM2-Smad4-depleted, and LM2-Smad4-Rescue cell lines were treated with 100 pM TGFβ for 3 h. TGFβ-induced fold changes of ANGPTL4 were analyzed by qRT-PCR analysis, n=3; error bars indicate standard deviation.
Figure 5. ANGPTL4 mediates TGFβ priming for mammary tumor dissemination to the lungs
(A) Secreted Angptl4 protein levels in the control, ANGPTL4 knockdown, or _ANGPTL4_-rescue LM2 cells were analyzed by ELISA. n=3; error bars indicate standard deviation (B) Tumors size measurements were taken from mice inoculated into the fourth mammary fat pads with 5×105 control, ANGPTL4 knockdown, or _ANGPTL4_-rescue LM2 cells. Tumor size was measured at day 28. n=14; error bars indicated s.e.m (C) The presence of circulating tumor cells was assessed by qRT-PCR as a function of human-specific GAPDH expression relative to murine β2-microglobulin, in 3 mL of mouse blood perfusate. n=10; error bars indicate s.e.m. (D) Peri-aortic lymph node metastasis from indicated tumors was analyzed by ex-vivo detection of luciferase activity. Positive bioluminescent signal in extracted lymph nodes indicated presence of metastasized tumor cells. (E) Photon flux measurements of breast cancer cells seeding the lung from orthotopically injected tumors as indicated. n=13–15 error bars indicated s.e.m; p-values calculated using the one-tailed unpaired t-test. (F) ANGPTL4 mRNA levels were determined by qRT-PCR analysis in cells that were incubated for 6 h with or without TGFβ. n=3; error bars indicate standard deviation. (G) Lung colonization analysis was performed by injecting 2×105 cells into the lateral tail vein. Prior to injection, cells were treated as indicated with 100 pM TGFβ for 6 h. Bar graphs represent 24 h time point measurements of the normalized photon flux. n=14–21; error bars indicate s.e.m; p-values calculated using the one-tailed unpaired t-test. (H) Normalized photon flux measurements from tail vein injected animals. Lung colonization measurements were taken from animals injected with control or Angptl4 overexpressing LM2 cells. n=6; error bars indicate s.e.m; p-values calculated using the one-tailed unpaired t-test.
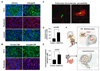
Figure 6. ANGPTL4 mediates endothelial monolayer disruption, lung capillary permeability, and trans-endothelial tumor cell migration
(A) HUVEC monolayers were grown to confluence on fibronectin coated slides and then treated for 24 h with rhAngptl4. Slides were subsequently fluorescently stained with anti-ZO-1 antibody, phalloidin, and anti-β-catenin antibody. (B) HUVEC monolayers were treated for 24 h with media conditioned by control LM2 cells or LM2 cells that overexpress Angptl4. Samples were stained for ZO-1 and phalloidin. (C) GFP-labeled MDA-231 cells were injected via the tail vein and allowed to lodge in the lungs. One day post injection, a rhodamine-dextran dye was injected into circulation. Three hours after dye injection, lungs were extracted and frozen sections were obtained. Representative confocal images are shown here of cells with and without accumulation of dye in the lung parenchyma. (D) Images were obtained as described in (C) with control or Angptl4 overexpressing MDA-MB-231 cells. A region of interest was drawn around the GFP-labeled cells and the amount of dextran dye was quantified based on rhodamine emissions. n=40 cells; error bars indicate s.e.m; p-values calculated using the one-tailed unpaired t-test. (E) Indicated cell lines were seeded into trans-well inserts that were previously covered with a HUVEC monolayer. Cells that migrated cross the endothelial layer into the bottom side of the transwell membrane were quantified with Volocity software. n=15, error bars indicate s.e.m; p-values calculated using the one-tailed unpaired t-test. (F) Schematic model of the cytokine relay set up by TGFβ activity in the primary tumor. ER− primary tumor cells that are exposed to TGFβ respond with ANGPTL4 induction via the Smad pathway. As they enter the circulation and reach the lung capillaries, these cells secrete Angptl4 which disrupts endothelial cell junctions thereby enabling the cancer cells to more efficiently enter the lung parenchyma.
Comment in
- TGFbeta primes breast tumor cells for metastasis.
Welm AL. Welm AL. Cell. 2008 Apr 4;133(1):27-8. doi: 10.1016/j.cell.2008.03.012. Cell. 2008. PMID: 18394984
Similar articles
- TGFbeta primes breast tumor cells for metastasis.
Welm AL. Welm AL. Cell. 2008 Apr 4;133(1):27-8. doi: 10.1016/j.cell.2008.03.012. Cell. 2008. PMID: 18394984 - TGFβ modulates inflammatory cytokines and growth factors to create premetastatic microenvironment and stimulate lung metastasis.
Ye Y, Liu S, Wu C, Sun Z. Ye Y, et al. J Mol Histol. 2015 Oct;46(4-5):365-75. doi: 10.1007/s10735-015-9633-4. Epub 2015 Jul 25. J Mol Histol. 2015. PMID: 26208571 - HIF-1-dependent expression of angiopoietin-like 4 and L1CAM mediates vascular metastasis of hypoxic breast cancer cells to the lungs.
Zhang H, Wong CC, Wei H, Gilkes DM, Korangath P, Chaturvedi P, Schito L, Chen J, Krishnamachary B, Winnard PT Jr, Raman V, Zhen L, Mitzner WA, Sukumar S, Semenza GL. Zhang H, et al. Oncogene. 2012 Apr 5;31(14):1757-70. doi: 10.1038/onc.2011.365. Epub 2011 Aug 22. Oncogene. 2012. PMID: 21860410 Free PMC article. Retracted. - Angiopoietin-like 4 prevents metastasis through inhibition of vascular permeability and tumor cell motility and invasiveness.
Galaup A, Cazes A, Le Jan S, Philippe J, Connault E, Le Coz E, Mekid H, Mir LM, Opolon P, Corvol P, Monnot C, Germain S. Galaup A, et al. Proc Natl Acad Sci U S A. 2006 Dec 5;103(49):18721-6. doi: 10.1073/pnas.0609025103. Epub 2006 Nov 27. Proc Natl Acad Sci U S A. 2006. PMID: 17130448 Free PMC article. - Transforming growth factor-beta signaling: emerging stem cell target in metastatic breast cancer?
Tan AR, Alexe G, Reiss M. Tan AR, et al. Breast Cancer Res Treat. 2009 Jun;115(3):453-95. doi: 10.1007/s10549-008-0184-1. Epub 2008 Oct 9. Breast Cancer Res Treat. 2009. PMID: 18841463 Free PMC article. Review.
Cited by
- T cell- but not tumor cell-produced TGF-β1 promotes the development of spontaneous mammary cancer.
Sarkar A, Donkor MK, Li MO. Sarkar A, et al. Oncotarget. 2011 Dec;2(12):1339-51. doi: 10.18632/oncotarget.403. Oncotarget. 2011. PMID: 22248703 Free PMC article. - The ELK3-GATA3 axis orchestrates invasion and metastasis of breast cancer cells in vitro and in vivo.
Kong SY, Kim KS, Kim J, Kim MK, Lee KH, Lee JY, Oh N, Park JI, Park JH, Heo SH, Shim SH, Lee DR, Kim KP, Park KS. Kong SY, et al. Oncotarget. 2016 Oct 4;7(40):65137-65146. doi: 10.18632/oncotarget.11427. Oncotarget. 2016. PMID: 27556500 Free PMC article. - Bone metastasis: the importance of the neighbourhood.
Croucher PI, McDonald MM, Martin TJ. Croucher PI, et al. Nat Rev Cancer. 2016 May 25;16(6):373-86. doi: 10.1038/nrc.2016.44. Nat Rev Cancer. 2016. PMID: 27220481 Review. - Cancer Cell-derived Secretory Factors in Breast Cancer-associated Lung Metastasis: Their Mechanism and Future Prospects.
Urooj T, Wasim B, Mushtaq S, Shah SNN, Shah M. Urooj T, et al. Curr Cancer Drug Targets. 2020;20(3):168-186. doi: 10.2174/1568009620666191220151856. Curr Cancer Drug Targets. 2020. PMID: 31858911 Free PMC article. Review. - Fish oil prevents breast cancer cell metastasis to bone.
Mandal CC, Ghosh-Choudhury T, Yoneda T, Choudhury GG, Ghosh-Choudhury N. Mandal CC, et al. Biochem Biophys Res Commun. 2010 Nov 26;402(4):602-7. doi: 10.1016/j.bbrc.2010.10.063. Epub 2010 Oct 28. Biochem Biophys Res Commun. 2010. PMID: 20971068 Free PMC article.
References
- Bernards R, Weinberg RA. A progression puzzle. Nature. 2002;418:823. - PubMed
- Bierie B, Moses HL. Tumour microenvironment: TGFbeta: the molecular Jekyll and Hyde of cancer. Nat Rev Cancer. 2006;6:506–520. - PubMed
- Bild AH, Potti A, Nevins JR. Linking oncogenic pathways with therapeutic opportunities. Nat Rev Cancer. 2006;6:735–741. - PubMed
- Brummelkamp TR, Bernards R, Agami R. Stable suppression of tumorigenicity by virus-mediated RNA interference. Cancer Cell. 2002;2:243–247. - PubMed
- Buck MB, Fritz P, Dippon J, Zugmaier G, Knabbe C. Prognostic significance of transforming growth factor beta receptor II in estrogen receptor-negative breast cancer patients. Clin Cancer Res. 2004;10:491–498. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- GM07739/GM/NIGMS NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- R37 CA034610-25/CA/NCI NIH HHS/United States
- P30 CA008748/CA/NCI NIH HHS/United States
- P01 CA094060/CA/NCI NIH HHS/United States
- P01 CA094060-01A10002/CA/NCI NIH HHS/United States
- T32 GM007739/GM/NIGMS NIH HHS/United States
- R37 CA034610/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials