Module map of stem cell genes guides creation of epithelial cancer stem cells - PubMed (original) (raw)
Module map of stem cell genes guides creation of epithelial cancer stem cells
David J Wong et al. Cell Stem Cell. 2008.
Abstract
Self-renewal is a hallmark of stem cells and cancer, but existence of a shared stemness program remains controversial. Here, we construct a gene module map to systematically relate transcriptional programs in embryonic stem cells (ESCs), adult tissue stem cells, and human cancers. This map reveals two predominant gene modules that distinguish ESCs and adult tissue stem cells. The ESC-like transcriptional program is activated in diverse human epithelial cancers and strongly predicts metastasis and death. c-Myc, but not other oncogenes, is sufficient to reactivate the ESC-like program in normal and cancer cells. In primary human keratinocytes transformed by Ras and I kappa B alpha, c-Myc increases the fraction of tumor-initiating cells by 150-fold, enabling tumor formation and serial propagation with as few as 500 cells. c-Myc-enhanced tumor initiation is cell-autonomous and independent of genomic instability. Thus, activation of an ESC-like transcriptional program in differentiated adult cells may induce pathologic self-renewal characteristic of cancer stem cells.
Figures
Figure 1. Gene Module Analysis of Stem Cells
(A) Flow chart of the steps in our gene module analysis of stem cells. (B) Mouse stem cell gene module map: a matrix of gene sets (rows) versus expression arrays (columns), where a red (or green) entry indicates that the individual genes within the gene set are significantly induced (or repressed) in the expression array more than expected by chance (FDR<0.05, p<0.01). The intensity of each entry corresponds to the average fold change of all of the genes within the gene set in the array. The arrays are labeled as differentiated cells or the type of stem cell. A subset of significant gene sets is shown; redundant gene sets were removed for clarity. Gene sets defined by gene ontology terms are listed in black; published gene sets defined by expression arrays of stem cells and differentiated cells are listed in purple; gene set defined by RNA interference experiments is listed in orange; gene set defined by genome-wide chromatin immunoprecipitation experiments is listed in blue. Note the clustering of two groups of stem cells, labeled “ESC-like” and “adult tissue stem”.
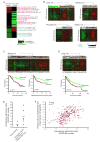
Figure 2. Activation of ESC Module in Human Cancers
(A) Cancer gene module map: a matrix of stem cell gene modules (columns) and array clinical annotations (rows), where a red (or green) entry indicates that the arrays in which the corresponding module was significantly induced (or repressed) contained more arrays with a given annotation than would be expected by chance (FDR<0.05, p<0.01). The intensity of the entries corresponds to the significance, i.e. −log10 (p-value). A subset of significant annotations is shown; redundant annotations were removed for clarity. (B-D) Expression patterns of ESC-like genes in primary human cancers and corresponding normal tissues are organized by hierarchical clustering. Note the coordinate regulation of the genes in the normal and cancer tissues. The dendrogram at the top of each data represent the similarities among the samples in their expression of ESC-like genes. Tumors are indicated by black branches, and normal tissues by green branches. Kaplan-Meier survival curves of tumors stratified into two classes based on expression of the ESC-like module are shown for data in (C) and (D). (E) Average log2 expression value of all the genes in the ESC-like module in previously published expression arrays of normal breast and FACS-sorted CD44+CD24-/low tumorigenic breast cancer cells (Liu et al., 2007). A line indicating mean expression value among all samples is shown for each. (F) Correlation of Pearson value to CD44+CD24-/low tumorigenic breast cancer cell signature with average log2 expression value of ESC-like module in the primary human breast cancers shown in (C).
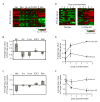
Figure 3. c-Myc Activates ESC Module in Epithelial Cells
(A) Expression of ESC-like module genes (excluding the proliferation genes) in previously published expression arrays of primary human mammary epithelial cells transduced with indicated oncogenes relative to those cells transduced with GFP (Bild et al., 2006). Each row represents an individual gene within the ESC-like module and each column is an expression array. The level of expression of each gene in each sample relative to the mean level of expression of that gene in the cells transduced with GFP is represented using a red-green color scale. (B) Average log2 expression values of all of the genes in the ESC-like gene module are expressed as mean ± SEM. (C) Average log2 expression values of all of the genes in the adult tissue stem gene module are expressed as mean ± SEM. (D) Expression of ESC-like module genes (excluding the proliferation genes) in previous published expression arrays of c-Myc induction in the epidermis of K14-Myc-ER transgenic mice, one and four days after topical application of tamoxifen (Frye et al., 2003). (E) Average log2 expression values of all of the genes in the ESC-like gene module are expressed as mean ± SEM. (F) Average log2 expression values of all of the genes in the adult tissue stem gene module are expressed as mean ± SEM.
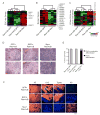
Figure 4. c-Myc Induces ESC-like State in Cancer
Subcutaneous scid mouse tumors derived from transduced primary human keratinocytes expressing the indicated proteins: (A-C) Each row represents an individual gene and each column is an expression array of a subcutaneous tumor. The level of expression of each gene in each sample was normalized relative to its average expression in all the samples and is represented using a red-green color scale. (A) RNA expression of genes in the ESC-like module. (B) RNA expression of genes with bivalent chromatin domains that are repressed in ESCs (Bernstein et al., 2006). (C) RNA expression of genes in a signature of CD44+CD24-/low cancer cell subpopulation enriched for cancer stem cells (Liu et al., 2007). The CD44+CD24-/low signature is composed of genes that are induced and repressed which is shown on the bar to the right (red is induced; green is repressed). (D) Histology of subcutaneous tumors. (E) Grades of subcutaneous tumors. (F) Subcutaneous tumor tissue: protein expression of keratinocyte differentiation markers keratin 5 (K5), keratin 10 (K10), transglutaminase 1 (Tgase), and keratin 8 (K8), all in orange. Hoechst 33342 nuclear staining in blue.
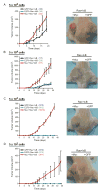
Figure 5. c-Myc Increases Fraction of Tumor-Initiating Cells
Left: Growth kinetics of tumors formed after subcutaneous injection of transduced primary human keratinocytes expressing the indicated proteins (mean ± SEM). Right: Representative pictures of subcutaneous tumors in scid mice: c-Myc+Ras+IκB tumors on left flank and GFP+Ras+IκB tumors on right flank. (A) 5×105 cells per subcutaneous injection. Picture is at day 18 post-injection. (B) 5×104 cells. The growth kinetics shown for GFP+Ras+IκB is of the one tumor that formed. The growth kinetics shown for E2F3+Ras+IκB is the mean ± SEM of the two tumors that formed. Picture is at day 20. (C) 5×103 cells. Picture is at day 28. (D) 5×102 cells. The growth kinetics shown for Myc+Ras+IκB is the mean ± SEM of the three tumors that formed. Picture is at day 45.
Figure 6. c-Myc-mediated Tumor Initation Is Cell-Autonomous and Not Linked with Genomic Instability
(A) Subcutaneous scid mouse tumors derived from injection of 5×104 transduced primary human keratinocytes expressing GFP, Ras, and IκB (100% GFP+Ras+IκB), 5×104 cells expressing c-Myc, Ras, and IκB (100% Myc+Ras+IκB), or 2.5×104 cells expressing GFP, Ras, and IκB plus 2.5×104 cells expressing c-Myc, Ras, and IκB (50% GFP+Ras+IκB, 50% Myc+Ras+IκB). Histology and immunohistochemistry with antibody against GFP. (B) Graphical displays of genome-wide DNA copy number alteration of subcutaneous tumors using array-based comparative genomic hybridization. Ratios are plotted on a log2 scale according to chromosome position.
Comment in
- Cancer: inappropriate expression of stem cell programs?
Wang Y, Armstrong SA. Wang Y, et al. Cell Stem Cell. 2008 Apr 10;2(4):297-9. doi: 10.1016/j.stem.2008.03.014. Cell Stem Cell. 2008. PMID: 18397746
Similar articles
- Cancer: inappropriate expression of stem cell programs?
Wang Y, Armstrong SA. Wang Y, et al. Cell Stem Cell. 2008 Apr 10;2(4):297-9. doi: 10.1016/j.stem.2008.03.014. Cell Stem Cell. 2008. PMID: 18397746 - Nac1 promotes self-renewal of embryonic stem cells through direct transcriptional regulation of c-Myc.
Ruan Y, He J, Wu W, He P, Tian Y, Xiao L, Liu G, Wang J, Cheng Y, Zhang S, Yang Y, Xiong J, Zhao K, Wan Y, Huang H, Zhang J, Jian R. Ruan Y, et al. Oncotarget. 2017 Jul 18;8(29):47607-47618. doi: 10.18632/oncotarget.17744. Oncotarget. 2017. PMID: 28548937 Free PMC article. - Transcriptional Regulator CNOT3 Defines an Aggressive Colorectal Cancer Subtype.
Cejas P, Cavazza A, Yandava CN, Moreno V, Horst D, Moreno-Rubio J, Burgos E, Mendiola M, Taing L, Goel A, Feliu J, Shivdasani RA. Cejas P, et al. Cancer Res. 2017 Feb 1;77(3):766-779. doi: 10.1158/0008-5472.CAN-16-1346. Epub 2016 Nov 29. Cancer Res. 2017. PMID: 27899379 Free PMC article. - Mechanisms that mediate stem cell self-renewal and differentiation.
Zhang H, Wang ZZ. Zhang H, et al. J Cell Biochem. 2008 Feb 15;103(3):709-18. doi: 10.1002/jcb.21460. J Cell Biochem. 2008. PMID: 17647265 Review. - Embryonic stem cell microRNAs: defining factors in induced pluripotent (iPS) and cancer (CSC) stem cells?
Gunaratne PH. Gunaratne PH. Curr Stem Cell Res Ther. 2009 Sep;4(3):168-77. doi: 10.2174/157488809789057400. Curr Stem Cell Res Ther. 2009. PMID: 19492978 Review.
Cited by
- Lineage factors and differentiation states in lung cancer progression.
Cheung WK, Nguyen DX. Cheung WK, et al. Oncogene. 2015 Nov 19;34(47):5771-80. doi: 10.1038/onc.2015.85. Epub 2015 Mar 30. Oncogene. 2015. PMID: 25823023 Free PMC article. Review. - Global levels of H3K27me3 track with differentiation in vivo and are deregulated by MYC in prostate cancer.
Pellakuru LG, Iwata T, Gurel B, Schultz D, Hicks J, Bethel C, Yegnasubramanian S, De Marzo AM. Pellakuru LG, et al. Am J Pathol. 2012 Aug;181(2):560-9. doi: 10.1016/j.ajpath.2012.04.021. Epub 2012 Jun 17. Am J Pathol. 2012. PMID: 22713676 Free PMC article. - The significance of PIWI family expression in human lung embryogenesis and non-small cell lung cancer.
Navarro A, Tejero R, Viñolas N, Cordeiro A, Marrades RM, Fuster D, Caritg O, Moises J, Muñoz C, Molins L, Ramirez J, Monzo M. Navarro A, et al. Oncotarget. 2015 Oct 13;6(31):31544-56. doi: 10.18632/oncotarget.3003. Oncotarget. 2015. PMID: 25742785 Free PMC article. - Functional genomics screening utilizing mutant mouse embryonic stem cells identifies novel radiation-response genes.
Loesch K, Galaviz S, Hamoui Z, Clanton R, Akabani G, Deveau M, DeJesus M, Ioerger T, Sacchettini JC, Wallis D. Loesch K, et al. PLoS One. 2015 Apr 8;10(4):e0120534. doi: 10.1371/journal.pone.0120534. eCollection 2015. PLoS One. 2015. PMID: 25853515 Free PMC article. - Global Profiling of the Lysine Crotonylome in Different Pluripotent States.
Lv Y, Bu C, Meng J, Ward C, Volpe G, Hu J, Jiang M, Guo L, Chen J, Esteban MA, Bao X, Cheng Z. Lv Y, et al. Genomics Proteomics Bioinformatics. 2021 Feb;19(1):80-93. doi: 10.1016/j.gpb.2021.01.004. Epub 2021 Mar 19. Genomics Proteomics Bioinformatics. 2021. PMID: 33746086 Free PMC article.
References
- Arnold I, Watt FM. c-Myc activation in transgenic mouse epidermis results in mobilization of stem cells and differentiation of their progeny. Curr Biol. 2001;11:558–568. - PubMed
- Arvanitis C, Felsher DW. Conditional transgenic models define how MYC initiates and maintains tumorigenesis. Semin Cancer Biol. 2006;16:313–317. - PubMed
- Beachy PA, Karhadkar SS, Berman DM. Tissue repair and stem cell renewal in carcinogenesis. Nature. 2004;432:324–331. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA119176/CA/NCI NIH HHS/United States
- R01-CA119176/CA/NCI NIH HHS/United States
- R01-CA118750/CA/NCI NIH HHS/United States
- R01 CA118750-02/CA/NCI NIH HHS/United States
- R01 CA118750/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources