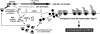Bacterial addiction module toxin Doc inhibits translation elongation through its association with the 30S ribosomal subunit - PubMed (original) (raw)
Bacterial addiction module toxin Doc inhibits translation elongation through its association with the 30S ribosomal subunit
Mohan Liu et al. Proc Natl Acad Sci U S A. 2008.
Abstract
Bacterial toxin-antitoxin (TA) systems (or "addiction modules") typically facilitate cell survival during intervals of stress by inducing a state of reversible growth arrest. However, upon prolonged stress, TA toxin action leads to cell death. TA systems have also been implicated in several clinically important phenomena: biofilm formation, bacterial persistence during antibiotic treatment, and bacterial pathogenesis. TA systems harbored by pathogens also serve as attractive antibiotic targets. To date, the mechanism of action of the majority of known TA toxins has not yet been elucidated. We determined the mode of action of the Doc toxin of the Phd-Doc TA system. Doc expression resulted in rapid cell growth arrest and marked inhibition of translation without significant perturbation of transcription or replication. However, Doc did not cleave mRNA as do other addiction-module toxins whose activities result in translation inhibition. Instead, Doc induction mimicked the effects of treatment with the aminoglycoside antibiotic hygromycin B (HygB): Both Doc and HygB interacted with 30S ribosomal subunits, stabilized polysomes, and resulted in a significant increase in mRNA half-life. HygB also competed with ribosome-bound Doc, whereas HygB-resistant mutants suppressed Doc toxicity, suggesting that the Doc-binding site includes that of HygB (i.e., helix 44 region of 16S rRNA containing the A, P, and E sites). Overall, our results illuminate an intracellular target and mechanism of TA toxin action drawn from aminoglycoside antibiotics: Doc toxicity is the result of inhibition of translation elongation, possibly at the translocation step, through its interaction with the 30S ribosomal subunit.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
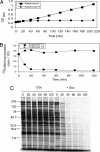
Doc expression leads to translation arrest. (A) Growth profile of Doc-induced (Arabinose+) or uninduced (Arabinose−) BW25113 cells containing pBAD33-Doc grown at 37°C in M9 medium containing 0.2% glycerol as the sole carbon source with 25 μg/ml chloramphenicol. (B) Quantification of [35S]methionine incorporation into Doc-induced (Arabinose+) or uninduced (Arabinose−) cells. (C) [35S]methionine incorporation of Doc-induced or uninduced cells in vivo. Equivalent amounts of cell lysate, derived from equal culture volumes, were subjected to SDS/PAGE, followed by autoradiography. Time points correspond to those in B; cell samples for experiments in B and C were derived from the growth-profile experiment shown in A. Molecular mass markers (kDa) are shown on the left.
Fig. 2.
Phd can rescue Doc-mediated in vitro translation arrest and in vivo growth arrest. (A) Purification of recombinant Phd-Doc–His6 complex after Ni-NTA-affinity chromatography (lane 1), followed by denaturation and refolding of purified Phd (lane 2), and recapture of Doc over Ni-NTA, followed by denaturation and refolding (lane 3). Molecular mass markers are shown on the left. (B) Recombinant Doc inhibits coupled in vitro transcription/translation (lane 3), Phd can rescue the inhibition and reconstitute translation (lane 2) to normal levels (as shown in lane 1); the product of the CAT fusion template (39 kDa) comprises one of the two major reaction products, the other β-lactamase (28 kDa) product is present at high levels in T7 S30 extracts because of transcription from the T7 promoter upstream of the CAT fusion that reads through into the ampicillin resistance gene. (C) Coexpression of the phd-doc module (upper right quadrant), doc alone (bottom quadrants), or empty pBAD33 plasmid (top left quadrant). The M9 plate containing 0.2% arabinose, 0.2% glycerol, and 25 μg/ml chloramphenicol was incubated at 37°C overnight.
Fig. 3.
Doc expression increases mRNA stability. RNA samples were prepared from rifampicin-treated+/− Doc cells; after Northern blot analysis of TufA and OmpA mRNAs, mRNA levels were then quantified and normalized to each uninduced time point.
Fig. 4.
Doc stabilizes polysomes, associates with 30S ribosomal subunits, and can be competed by HygB. (A) −HygB −Doc ribosomes were prepared from BW25113 cells containing pBAD33-Doc under noninducing conditions with no HygB added. (B) +HygB −Doc, BW25113 cells containing pBAD33-Doc were treated with 50 μg/ml HygB only for 10 min. (C) −HygB +Doc, BW25113 cells containing pBAD33-Doc were induced with 0.2% arabinose only for 20 min. (D) Fractions corresponding to free 30S and 50S ribosomal subunits, 70S monosomes, and polysomes from samples treated as labeled above the graphs were collected and subjected to Western blot analysis with a Doc polyclonal antibody. Samples for the HygB competition experiments in lanes 4–6 were exposed to 50 μg/ml, 200 μg/ml, or 500 μg/ml HygB before cell harvesting and ribosome fractionation. We observed strong chemiluminescence with very short exposure times; high-intensity bands appear with varying degrees of white centers that diminish with increasing concentration of HygB exposure. Because the two −Doc polysome samples were not loaded adjacent to the four +Doc polysome samples, they were digitally placed in the same order as the other frames in D.
Fig. 5.
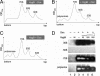
Doc is not toxic in a HygB-resistant mutant. (A and B) S30 extracts were prepared from E. coli cells containing pKK1491U and pBAD33-Doc. Results shown were confirmed in two independent experiments. (C) Comparison of plate growth phenotypes of BW25113 cells containing pBAD33-Doc and pKK1491U to BW25113 wild-type cells containing only pBAD33-Doc subjected to the conditions shown. Relative growth rates (strong/wild type (++++), intermediate (+++) weak (+/−), or no (−) growth) were scored after viewing plates grown for the same amount of time overnight in LB plus relevant selective antibiotic at 37°C.
Fig. 6.
Features of Phd-Doc function in E. coli. Toxicity occurs when free Doc toxin is able to arrest translation elongation by binding to the 30S subunit (denoted by the X); unchecked toxin action leads to bacterial cell death (i.e. postsegregational killing for P1 bacteriophage infected cells). The mechanism of mRNA stabilization by Doc can also be envisioned because stalled ribosomes protect mRNA from degradation. Note that all 30S subunits do not necessarily have to be bound by Doc for both mRNA stabilization and translation arrest to occur because stalled ribosomes block read-through of ribosomes before them.
Similar articles
- Bacterial toxin YafQ is an endoribonuclease that associates with the ribosome and blocks translation elongation through sequence-specific and frame-dependent mRNA cleavage.
Prysak MH, Mozdzierz CJ, Cook AM, Zhu L, Zhang Y, Inouye M, Woychik NA. Prysak MH, et al. Mol Microbiol. 2009 Mar;71(5):1071-87. doi: 10.1111/j.1365-2958.2008.06572.x. Epub 2009 Jan 30. Mol Microbiol. 2009. PMID: 19210620 - Doc toxin is a kinase that inactivates elongation factor Tu.
Cruz JW, Rothenbacher FP, Maehigashi T, Lane WS, Dunham CM, Woychik NA. Cruz JW, et al. J Biol Chem. 2014 Mar 14;289(11):7788-98. doi: 10.1074/jbc.M113.544429. Epub 2014 Jan 21. J Biol Chem. 2014. PMID: 24448800 Free PMC article. - Doc of prophage P1 is inhibited by its antitoxin partner Phd through fold complementation.
Garcia-Pino A, Christensen-Dalsgaard M, Wyns L, Yarmolinsky M, Magnuson RD, Gerdes K, Loris R. Garcia-Pino A, et al. J Biol Chem. 2008 Nov 7;283(45):30821-7. doi: 10.1074/jbc.M805654200. Epub 2008 Aug 30. J Biol Chem. 2008. PMID: 18757857 Free PMC article. - Bacterial toxin-antitoxin modules: classification, functions, and association with persistence.
Singh G, Yadav M, Ghosh C, Rathore JS. Singh G, et al. Curr Res Microb Sci. 2021 Jul 7;2:100047. doi: 10.1016/j.crmicr.2021.100047. eCollection 2021 Dec. Curr Res Microb Sci. 2021. PMID: 34841338 Free PMC article. Review. - Ribonucleases in bacterial toxin-antitoxin systems.
Cook GM, Robson JR, Frampton RA, McKenzie J, Przybilski R, Fineran PC, Arcus VL. Cook GM, et al. Biochim Biophys Acta. 2013 Jun-Jul;1829(6-7):523-31. doi: 10.1016/j.bbagrm.2013.02.007. Epub 2013 Feb 21. Biochim Biophys Acta. 2013. PMID: 23454553 Review.
Cited by
- Discovery of functional toxin/antitoxin systems in bacteria by shotgun cloning.
Sberro H, Leavitt A, Kiro R, Koh E, Peleg Y, Qimron U, Sorek R. Sberro H, et al. Mol Cell. 2013 Apr 11;50(1):136-48. doi: 10.1016/j.molcel.2013.02.002. Epub 2013 Mar 7. Mol Cell. 2013. PMID: 23478446 Free PMC article. - Crystal structure of the VapBC toxin-antitoxin complex from Shigella flexneri reveals a hetero-octameric DNA-binding assembly.
Dienemann C, Bøggild A, Winther KS, Gerdes K, Brodersen DE. Dienemann C, et al. J Mol Biol. 2011 Dec 16;414(5):713-22. doi: 10.1016/j.jmb.2011.10.024. Epub 2011 Oct 20. J Mol Biol. 2011. PMID: 22037005 Free PMC article. - Bacterial toxin-antitoxin systems: more than selfish entities?
Van Melderen L, Saavedra De Bast M. Van Melderen L, et al. PLoS Genet. 2009 Mar;5(3):e1000437. doi: 10.1371/journal.pgen.1000437. Epub 2009 Mar 27. PLoS Genet. 2009. PMID: 19325885 Free PMC article. Review. - AMPylation: Something Old is New Again.
Woolery AR, Luong P, Broberg CA, Orth K. Woolery AR, et al. Front Microbiol. 2010 Oct 19;1:113. doi: 10.3389/fmicb.2010.00113. eCollection 2010. Front Microbiol. 2010. PMID: 21607083 Free PMC article. - Mycobacterium tuberculosis Type II Toxin-Antitoxin Systems: Genetic Polymorphisms and Functional Properties and the Possibility of Their Use for Genotyping.
Zaychikova MV, Zakharevich NV, Sagaidak MO, Bogolubova NA, Smirnova TG, Andreevskaya SN, Larionova EE, Alekseeva MG, Chernousova LN, Danilenko VN. Zaychikova MV, et al. PLoS One. 2015 Dec 14;10(12):e0143682. doi: 10.1371/journal.pone.0143682. eCollection 2015. PLoS One. 2015. PMID: 26658274 Free PMC article.
References
- Gerdes K, Christensen SK, Lobner-Olesen A. Prokaryotic toxin–antitoxin stress response loci. Nat Rev Microbiol. 2005;3:371–382. - PubMed
- Suzuki M, Zhang J, Liu M, Woychik NA, Inouye M. Single protein production in living cells facilitated by an mRNA interferase. Mol Cell. 2005;18:253–261. - PubMed
- Engelberg-Kulka H, Hazan R, Amitai S. mazEF: A chromosomal toxin–antitoxin module that triggers programmed cell death in bacteria. J Cell Sci. 2005;118:4327–4332. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases