Detection and prevalence patterns of group I coronaviruses in bats, northern Germany - PubMed (original) (raw)
doi: 10.3201/eid1404.071439.
Anne Ipsen, Antje Seebens, Matthias Göttsche, Marcus Panning, Jan Felix Drexler, Nadine Petersen, Augustina Annan, Klaus Grywna, Marcel Müller, Susanne Pfefferle, Christian Drosten
Affiliations
- PMID: 18400147
- PMCID: PMC2570906
- DOI: 10.3201/eid1404.071439
Detection and prevalence patterns of group I coronaviruses in bats, northern Germany
Florian Gloza-Rausch et al. Emerg Infect Dis. 2008 Apr.
Abstract
We tested 315 bats from 7 different bat species in northern Germany for coronaviruses by reverse transcription-PCR. The overall prevalence was 9.8%. There were 4 lineages of group I coronaviruses in association with 4 different species of verspertilionid bats (Myotis dasycneme, M. daubentonii, Pipistrellus nathusii, P. pygmaeus). The lineages formed a monophyletic clade of bat coronaviruses found in northern Germany. The clade of bat coronaviruses have a sister relationship with a clade of Chinese type I coronaviruses that were also associated with the Myotis genus (M. ricketti). Young age and ongoing lactation, but not sex or existing gravidity, correlated significantly with coronavirus detection. The virus is probably maintained on the population level by amplification and transmission in maternity colonies, rather than being maintained in individual bats.
Figures
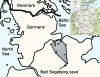
Figure 1
Map of Germany (inset) with enlarged view of northern Germany. The study area is shaded, and dots in the study area indicate sampling sites.
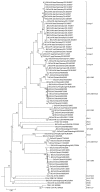
Figure 2
Phlyogenetic analysis of northern German bat coronaviruses (CoV) (lineages 1–4) and related group I CoVs from bats and other mammals. Analyses were conducted in MEGA4 (32), by using the neighbor-joining algorithm with Kimura correction and a bootstrap test of phylogeny. Numbers at nodes denote bootstrap values as percentage of 1,000 repetitive analyses. The phylogeny is rooted with a Leopard CoV, ALC/GX/F230/06 (33). The column on the right shows bat CoV prototype strain names or the designations of type strains of established mammalian CoV species.
Similar articles
- Detection of group 1 coronaviruses in bats in North America.
Dominguez SR, O'Shea TJ, Oko LM, Holmes KV. Dominguez SR, et al. Emerg Infect Dis. 2007 Sep;13(9):1295-300. doi: 10.3201/eid1309.070491. Emerg Infect Dis. 2007. PMID: 18252098 Free PMC article. - Circulation of group 2 coronaviruses in a bat species common to urban areas in Western Europe.
Reusken CB, Lina PH, Pielaat A, de Vries A, Dam-Deisz C, Adema J, Drexler JF, Drosten C, Kooi EA. Reusken CB, et al. Vector Borne Zoonotic Dis. 2010 Oct;10(8):785-91. doi: 10.1089/vbz.2009.0173. Epub 2010 Jan 7. Vector Borne Zoonotic Dis. 2010. PMID: 20055576 - Coronaviruses (Coronaviridae) of bats in the northern Caucasus and south of western Siberia.
Yashina LN, Zhigalin AV, Abramov SA, Luchnikova EM, Smetannikova NA, Dupal TA, Krivopalov AV, Vdovina ED, Svirin KA, Gadzhiev AA, Malyshev BS. Yashina LN, et al. Vopr Virusol. 2024 Jul 5;69(3):255-265. doi: 10.36233//0507-4088-233. Vopr Virusol. 2024. PMID: 38996374 - Coronaviruses in Bats: A Review for the Americas.
Hernández-Aguilar I, Lorenzo C, Santos-Moreno A, Naranjo EJ, Navarrete-Gutiérrez D. Hernández-Aguilar I, et al. Viruses. 2021 Jun 25;13(7):1226. doi: 10.3390/v13071226. Viruses. 2021. PMID: 34201926 Free PMC article. Review. - Bat Coronaviruses in China.
Fan Y, Zhao K, Shi ZL, Zhou P. Fan Y, et al. Viruses. 2019 Mar 2;11(3):210. doi: 10.3390/v11030210. Viruses. 2019. PMID: 30832341 Free PMC article. Review.
Cited by
- Parasite diversity of European Myotis species with special emphasis on Myotis myotis (Microchiroptera, Vespertilionidae) from a typical nursery roost.
Frank R, Kuhn T, Werblow A, Liston A, Kochmann J, Klimpel S. Frank R, et al. Parasit Vectors. 2015 Feb 15;8:101. doi: 10.1186/s13071-015-0707-7. Parasit Vectors. 2015. PMID: 25880235 Free PMC article. - Coronaviruses Are Abundant and Genetically Diverse in West and Central African Bats, including Viruses Closely Related to Human Coronaviruses.
Meta Djomsi D, Lacroix A, Soumah AK, Kinganda Lusamaki E, Mesdour A, Raulino R, Esteban A, Ndong Bass I, Mba Djonzo FA, Goumou S, Ndimbo-Kimugu SP, Lempu G, Mbala Kingebeni P, Bamuleka DM, Likofata J, Muyembe Tamfum JJ, Toure A, Mpoudi Ngole E, Kouanfack C, Delaporte E, Keita AK, Ahuka-Mundeke S, Ayouba A, Peeters M. Meta Djomsi D, et al. Viruses. 2023 Jan 25;15(2):337. doi: 10.3390/v15020337. Viruses. 2023. PMID: 36851551 Free PMC article. - Detection and Characterization of Distinct Alphacoronaviruses in Five Different Bat Species in Denmark.
Lazov CM, Chriél M, Baagøe HJ, Fjederholt E, Deng Y, Kooi EA, Belsham GJ, Bøtner A, Rasmussen TB. Lazov CM, et al. Viruses. 2018 Sep 11;10(9):486. doi: 10.3390/v10090486. Viruses. 2018. PMID: 30208582 Free PMC article. - Identification and characterization of novel alphacoronaviruses in Tadarida brasiliensis (Chiroptera, Molossidae) from Argentina: insights into recombination as a mechanism favoring bat coronavirus cross-species transmission.
Cerri A, Bolatti EM, Zorec TM, Montani ME, Rimondi A, Hosnjak L, Casal PE, Di Domenica V, Barquez RM, Poljak M, Giri AA. Cerri A, et al. Microbiol Spectr. 2023 Sep 11;11(5):e0204723. doi: 10.1128/spectrum.02047-23. Online ahead of print. Microbiol Spectr. 2023. PMID: 37695063 Free PMC article. - Antibodies against MERS coronavirus in dromedary camels, United Arab Emirates, 2003 and 2013.
Meyer B, Müller MA, Corman VM, Reusken CB, Ritz D, Godeke GJ, Lattwein E, Kallies S, Siemens A, van Beek J, Drexler JF, Muth D, Bosch BJ, Wernery U, Koopmans MP, Wernery R, Drosten C. Meyer B, et al. Emerg Infect Dis. 2014 Apr;20(4):552-9. doi: 10.3201/eid2004.131746. Emerg Infect Dis. 2014. PMID: 24655412 Free PMC article.
References
- Cavanagh D. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch Virol. 1997;142:629–33. - PubMed
- Hamre D, Procknow JJ. A new virus isolated from the human respiratory tract. Proc Soc Exp Biol Med. 1966;121:190–3. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources