The FUSE binding protein is a cellular factor required for efficient replication of hepatitis C virus - PubMed (original) (raw)
The FUSE binding protein is a cellular factor required for efficient replication of hepatitis C virus
Zhengbin Zhang et al. J Virol. 2008 Jun.
Abstract
Hepatitis C virus (HCV) infection is the leading cause of liver cirrhosis and hepatocellular carcinoma and one of the primary indications for liver transplantation. The molecular mechanisms underlying the actions of host factors in HCV replication remain poorly defined. FUSE (far upstream element of the c-myc proto-oncogene) binding protein (FBP) is a cellular factor that we have identified as a binder of HCV 3' nontranslated region (3'NTR). Mapping of the binding site showed that FBP specifically interacts with the poly(U) tract within the poly(U/UC) region of the 3'NTR. Silencing of FBP expression by small interfering RNA in cells carrying HCV subgenomic replicons severely reduced viral replication, while overexpression of FBP significantly enhanced viral replication. We confirmed these observations by an in vitro HCV replication assay in the cell-free replicative lysate, which suggested that there is a direct correlation between the cellular FBP level and HCV replication. FBP immunoprecipitation coprecipitated HCV nonstructural protein 5A (NS5A), indicating that FBP interacts with HCV NS5A, which is known to function as a link between HCV translation and replication. Although FBP is mainly localized in the nucleus, we found that in MH14 cells a significant level of this protein is colocalized with NS5A in the cytosol, a site of HCV replication. While the mechanism of FBP involvement in HCV replication is yet to be delineated, our findings suggest that it may be an important regulatory component that is essential for efficient replication of HCV.
Figures
FIG. 1.
Interaction between cytoplasmic FBP and Cy5-labeled HCV 3′NTR. Cy5-labeled, in vitro-transcribed HCV 3′NTR RNA was incubated with cytoplasmic extract from cured MH14 cells for 20 min at 37°C and then UV irradiated. The sample was then treated with RNase A and resolved by SDS-PAGE. The cross-linked (X-link) protein-RNA complexes were detected by using a Typhoon scanner (lane 1). We then transferred the RNA-protein complexes to the nitrocellulose membrane and Western blotted (WB) using anti-FBP antibody (lane 2). The positions of molecular mass protein markers (in kilodaltons) are shown to the left of the gel.
FIG. 2.
Specificity of interaction between FBP and HCV 3′NTR. (A) Cross-linking of Cy5-labeled HCV 3′NTR (HCV 3′NTR-Cy5) with FBP as a function of increasing FBP concentration. Cy5-labeled 3′NTR (0.5 pmol) was incubated with 0.25, 0.5, and 1.0 pmol of purified FBP (lanes 1 through 3, respectively). Lane 4 contained 0.5 pmol of purified FBP only. The samples were UV irradiated, then treated with RNase A, and resolved by SDS-PAGE. The cross-linked RNA-protein complexes were detected with a Typhoon scanner. (B) FBP cross-linking with 3′NTR as a function of the concentration of Cy5-labeled HCV 3′NTR and competition by increasing concentrations of cold HCV 3′NTR. In lanes 1 through 3, a fixed concentration of FBP (0.5 pmol) was cross-linked with 0.5, 0.25, and 0.125 pmol of Cy5-labeled HCV 3′NTR, respectively. In lanes 4 though 6, a fixed concentration of FBP (0.5 pmol) was cross-linked with a fixed concentration of Cy5-labeled HCV 3′NTR (0.5 pmol) in the presence of 0.5, 2.5, and 5 pmol of cold HCV 3′NTR, respectively. Lane 7 contained Cy5-labeled HCV 3′NTR (0.05 pmol) only. (C) FBP cross-linking with HCV 3′NTR in the presence of nonspecific heteropolymeric HIV-1 TAR RNA as the competitor. Lane 1 contains no competitor. Lane 2 contains 5 pmol of cold HIV-1 TAR RNA. Lane 3 contains 5 pmol of cold HCV 3′NTR. (D) FBP cross-linking with Cy5-labeled HCV 3′NTR in the presence of poly(rU) or poly(rC) as the competitor. Lane 1 contains no competitor, while lanes 2 and 3 contain poly(rU) and poly(rC) as the competitor, respectively. The positions of molecular mass markers (in kilodaltons) are shown to the left of the gels.
FIG. 3.
FBP binds to the poly(U/UC) region of HCV 3′NTR. (A) Schematic representation of PCR amplification and in vitro transcription of full-length HCV 3′NTR (F13) and its fragments corresponding to the variable region (F11), poly(U/UC) region (F22), and conserved 3′ X-tail (F33). (B) Cross-linking of FBP with Cy5-labeled full-length HCV 3′NTR (3′NTR-Cy5) (F13) in the absence (−) and presence (+) of in vitro-transcribed cold RNA fragments corresponding to F11, F22, and F33. Lane 1, 0.5 pmol of Cy5-labeled full-length HCV 3′NTR RNA (F13) alone without FBP; lane 2, 0.5 pmol of purified FBP cross-linked with 0.5 pmol of Cy5-labeled HCV 3′NTR in the absence of competitor. Lanes 3 through 5 show cross-linking of FBP with 0.5 pmol of Cy5-labeled HCV 3′NTR (F13) in the presence of a 10-fold excess of competitor RNA corresponding to F11, F22, and F33, respectively. The positions of molecular mass markers (in kilodaltons) are shown to the left of the gel. (C) Direct cross-linking of FBP with Cy5-labeled F11, F22, F33, or F13. Purified FBP (0.5 pmol) was cross-linked with 0.5 pmol of Cy5-labeled RNA fragments corresponding to F11, F22, F33, and full-length HCV 3′NTR (F13). The individual Cy5-labeled input RNA probe is shown in the bottom blot.
FIG. 4.
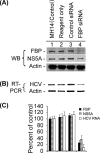
Effect of siRNA-mediated reduced expression of FBP on HCV replication. MH14 cells harboring replicating subgenomic HCV replicons were transfected with FBP siRNA. At 48 h posttransfection, the cells were harvested, lysed, and Western blotted (WB) for FBP, NS5A, and actin (A) and subjected to RT-PCR for HCV RNA and actin mRNA (B). (C) Quantification of WB and RT-PCR results.
FIG. 5.
Effects of down-regulation and up-regulation of FBP on the translation of Luc-HCV RNA replicon in cured MH14 cells. Cured MH14 cells transfected with either FBP siRNA or pCIA-CMV-FBP were grown for 24 h and then transfected with Luc-HCV RNA containing the luciferase reporter gene (Luc). At 24 h posttransfection, cells were harvested, lysed, and Western blotted (WB) for FBP and actin (A) and the results were quantified (B). An aliquot of the lysate was assayed for luciferase (Luc) activity (C).
FIG. 6.
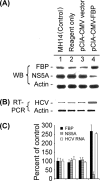
Effect of overexpression of FBP on HCV replication in MH14 cells. After 48 h of transfection with the FBP-expressing plasmid pCIA-CMV-FBP, MH14 cells were harvested, lysed, and Western blotted (WB) for FBP, NS5A, and actin (A) and then subjected to RT-PCR for HCV RNA and actin mRNA (B). (C) Quantification of WB and RT-PCR results.
FIG. 7.
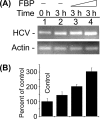
Stimulation of in vitro HCV replication in cell-free replicative lysate of MH14 cells by the addition of purified FBP. An aliquot of cell-free replicative lysate of MH14 cells was incubated in the absence (−) and presence of 0.25 and 0.5 pmol of FBP under standard reaction conditions. At the end of incubation, RT-PCR for HCV RNA (A) was done. (B) Quantification of HCV RNA results.
FIG. 8.
Coimmunoprecipitation of FBP and NS5A. (A) MH14 cell lysate was treated with RNase A and then incubated with anti-FBP antibody and protein A/G Plus agarose beads overnight at 4°C to immobilize the FBP-FBP antibody complex on the beads. The beads were washed several times, and the bound protein complex was resolved by SDS-PAGE, transferred onto the membrane, and Western blotted (WB) for NS5A using anti-NS5A antibody (lane 1). The lane with beads only (without FBP antibody) was used as a negative control (lane 2). The crude cell lysate from MH14 cell was used as a positive control (lane 3). FBP IP, FBP immunoprecipitate. (B) The protein bands in the immunoprecipitates were resolved on SDS-polyacrylamide gels and visualized by Coomassie blue staining. The positions of molecular mass markers (M) (in kilodaltons) are shown to the left of the gels in panels A and B. IgG, immunoglobulin G. RNase A-mediated degradation of cellular 28S and 18S RNAs and HCV RNA in MH14 cell lysate under the experimental conditions is shown in panels C and D, respectively.
FIG. 9.
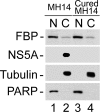
Cellular distribution of FBP and NS5A in MH14 cells. Nuclear (N) and cytoplasmic (C) extracts from MH14 and cured MH14 cells were prepared and Western blotted for FBP and NS5A. PARP and α-tubulin were also Western blotted as the specific nuclear and cytosolic markers, respectively.
FIG. 10.
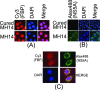
Cellular localization of FBP and NS5A in MH14 cells. (A) Localization of FBP in MH14 cells and cured MH14 cells. Cells were grown on a coverslip for 24 h, fixed, treated with anti-FBP antibody, and then treated with Cy3-labeled secondary antibody. DAPI was used to stain the nuclei. (B) Localization of HCV NS5A in MH14 cells and cured MH14 cells (negative control). Cells were fixed and treated with anti-NS5A antibody and then treated with Alexa Fluor 488 (Alex488)-labeled secondary antibody. DAPI was used to stain the nuclei. (C) Colocalization of FBP and NS5A in cytosol in MH14 cells. MH14 cells were fixed, treated with anti-FBP antibody and Cy3-labeled secondary antibody, and then treated with anti-NS5A antibody and Alexa Fluor 488-labeled secondary antibody. DAPI was used to stain the nuclei. They were then individually observed for FBP and NS5A localization. The same slide and the same field in different channels were used to observe the DAPI-stained nucleus, Cy3-labeled FBP, and Alexa 488-labeled NS5A. Pictures were taken and merged using Spot advanced software (Diagnostic Instruments, Sterling Heights, MI).
FIG. 11.
Effect of down- or up-regulation of FBP on c-myc expression in MH14 cells. (A) FBP expression in MH14 cells was either reduced by transfection of FBP siRNA (lanes 1 to 4) or enhanced by transfection of pCIA-CMV-FBP (lanes 5 to 8). At 48 h posttransfection, the cells were lysed and Western blotted for FBP and c-myc. (B) Quantification of the Western blot of FBP and c-myc proteins.
FIG. 12.
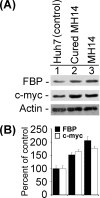
Expression levels of FBP and c-myc proteins in MH14 cells and cured MH14 cells and their parental Huh7 cells. The equivalent protein from cell lysates of MH14 cells, cured MH14 cells, and Huh7 cells were Western blotted for FBP and c-myc proteins (A), and the results were quantified (B).
Similar articles
- Y-Box Binding Protein 1 Stabilizes Hepatitis C Virus NS5A via Phosphorylation-Mediated Interaction with NS5A To Regulate Viral Propagation.
Wang WT, Tsai TY, Chao CH, Lai BY, Wu Lee YH. Wang WT, et al. J Virol. 2015 Nov;89(22):11584-602. doi: 10.1128/JVI.01513-15. Epub 2015 Sep 9. J Virol. 2015. PMID: 26355086 Free PMC article. - Nonstructural 5A protein activates beta-catenin signaling cascades: implication of hepatitis C virus-induced liver pathogenesis.
Park CY, Choi SH, Kang SM, Kang JI, Ahn BY, Kim H, Jung G, Choi KY, Hwang SB. Park CY, et al. J Hepatol. 2009 Nov;51(5):853-64. doi: 10.1016/j.jhep.2009.06.026. Epub 2009 Aug 12. J Hepatol. 2009. PMID: 19726098 - Dominant negative effect of wild-type NS5A on NS5A-adapted subgenomic hepatitis C virus RNA replicon.
Graziani R, Paonessa G. Graziani R, et al. J Gen Virol. 2004 Jul;85(Pt 7):1867-1875. doi: 10.1099/vir.0.80006-0. J Gen Virol. 2004. PMID: 15218171 - Phosphorylation of hepatitis C virus NS5A nonstructural protein: a new paradigm for phosphorylation-dependent viral RNA replication?
Huang Y, Staschke K, De Francesco R, Tan SL. Huang Y, et al. Virology. 2007 Jul 20;364(1):1-9. doi: 10.1016/j.virol.2007.01.042. Epub 2007 Apr 2. Virology. 2007. PMID: 17400273 Review. - Hepatitis C virus NS5A: tales of a promiscuous protein.
Macdonald A, Harris M. Macdonald A, et al. J Gen Virol. 2004 Sep;85(Pt 9):2485-2502. doi: 10.1099/vir.0.80204-0. J Gen Virol. 2004. PMID: 15302943 Review.
Cited by
- Comparative structural analyses and nucleotide-binding characterization of the four KH domains of FUBP1.
Ni X, Knapp S, Chaikuad A. Ni X, et al. Sci Rep. 2020 Aug 10;10(1):13459. doi: 10.1038/s41598-020-69832-z. Sci Rep. 2020. PMID: 32778776 Free PMC article. - Update on the Development of Anti-Viral Agents Against Hepatitis C.
Macarthur KL, Smolic R, Smolic MV, Wu CH, Wu GY. Macarthur KL, et al. J Clin Transl Hepatol. 2013 Sep;1(1):9-21. doi: 10.14218/JCTH.2013.007XX. Epub 2013 Sep 15. J Clin Transl Hepatol. 2013. PMID: 26357602 Free PMC article. Review. - Far upstream element-binding protein 1 and RNA secondary structure both mediate second-step splicing repression.
Li H, Wang Z, Zhou X, Cheng Y, Xie Z, Manley JL, Feng Y. Li H, et al. Proc Natl Acad Sci U S A. 2013 Jul 16;110(29):E2687-95. doi: 10.1073/pnas.1310607110. Epub 2013 Jul 1. Proc Natl Acad Sci U S A. 2013. PMID: 23818605 Free PMC article. - FUSE binding protein 1 interacts with untranslated regions of Japanese encephalitis virus RNA and negatively regulates viral replication.
Chien HL, Liao CL, Lin YL. Chien HL, et al. J Virol. 2011 May;85(10):4698-706. doi: 10.1128/JVI.01950-10. Epub 2011 Mar 2. J Virol. 2011. PMID: 21367899 Free PMC article.
References
- Abou-Elella, A., T. Gramlich, C. Fritsch, and T. Gansler. 1996. c-myc amplification in hepatocellular carcinoma predicts unfavorable prognosis. Mod. Pathol. 995-98. - PubMed
- Anwar, A., N. Ali, R. Tanveer, and A. Siddiqui. 2000. Demonstration of functional requirement of polypyrimidine tract-binding protein by SELEX RNA during hepatitis C virus internal ribosome entry site-mediated translation initiation. J. Biol. Chem. 27534231-34235. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous