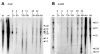Zebrafish brd2a and brd2b are paralogous members of the bromodomain-ET (BET) family of transcriptional coregulators that show structural and expression divergence - PubMed (original) (raw)
Figure 1
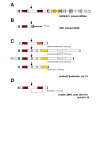
_brd2_-related genes encode predicted proteins with varying domain configurations. Schematics of cloned and predicted _brd2_-related cDNAs with encoded protein domains are shown for A) cloned zf626 and zf619 cDNAs, B) cloned zf69 cDNA, C) supported and predicted simbrd2 transcripts from chromosome 16 ENSDARG00000046087 locus from Ensembl and NCBI databases, and D) brd2b partial cDNA from NCBI database. Some schematics include 5'UTR and 3' UTR, with wider rectangles depicting protein-coding regions. Boxes show conserved domains: bromodomain (red box); nuclear localization signal (horizontal striped); ET domain (yellow box); in C, degenerate second bromodomain (orange box); in C, upstream TAP domain (light blue box); valine-, leucine-rich region (VL-rich box); arginine-, serine-rich region (RS-rich box); start and stop codons (thick vertical lines); in A, SNPs between zf626 and zf619 (thin vertical lines); in B, discontinuous region in zf69 cDNA with repetitive sequences at bases 921–1033 and 1259–1413 (gray box); conserved intron/exon junction for alternative splicing in _brd2_-related transcripts (arrow). Two-exon region in brd2b cDNA found in scaf_NA1181 is underscored in D. Length of cDNAs in base pairs is given in parentheses.
Figure 9
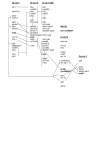
Syntenic regions shared by zebrafish chromosomes 19 and 16, human chromosome 6, and trout chromosome 2. Gene content of genomic regions where _brd2_-related sequences reside on human chromosome 6 (Hs chr 6), zebrafish chromosomes 19 (Dr chr 19) and 16 (Dr chr 16) from the Tubingen strain, zebrafish chromosome 19 from the AB strain (Dr chr 19/AB), and Onchorynchus chromosome 2 (Om chr 2). Gene order is derived from contig views in Vega (Dr chr 19, Dr chr 19/AB), Ensembl (Dr chr 16, Hs chr 6, NA1181), direct reading of annotated genomic clones (Dr chr 19, BX510994.6; Dr chr 19/AB, AL672176.10), and reference 28 (Om chr 2). Brd2-related genes are shown in bold. Genes that reside on the same genomic clone are shown in single-spaced groups; genomic clones within the same chromosome that physically overlap are connected by vertical lines. Related genes or groups of genes between chromosomes that show conserved synteny are also connected by lines. Human brd2 resides within the MHC II complex on chromosome 6, in a conserved syntenic block between psmb8 and rxrb genes; zebrafish brd2 is found in the same gene block within the fish MHC I core on chromosome 19, and is the true ortholog of mammalian brd2. Gene content on chromosome 19 from strains Tubingen and AB differs outside the psmb8-rxrb and the flot1-tpsn gene blocks, and may indicate recent genomic rearrangement. The zebrafish brd2b paralogous locus on chromosome 16 (alias loc 569354, sim brd2) is linked to tap1 sequences and other MHC II and III related genes, as is trout brd2L on chromosome 2, presumably representing the ancestral vertebrate configuration. sim loc569354 on unmapped scaffold NA1181 consists of two exons encoding the second bromodomain of brd2b partial cDNA (GenBank: BC055508), and with further genome assembly will likely be shown to derive from the same brd2b locus on chromosome 16.
Figure 2
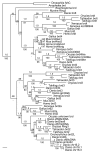
brd2 and simbrd2/brd2b represent a paralogous gene set in zebrafish. Nucleic acid maximum likelihood tree of selected BET family homologs with Bayesian posterior probabilities (upper) and 1000× parsimony bootstrap values (lower) added to branches. At least one duplication of the brd2 gene has occurred in teleosts. zf626/zf619 cDNAs group with brd2 in zebrafish, while zf69 cDNA groups with sequences from the simbrd2/brd2b paralogous locus. Danio zf626, GenBank: EU126947; Danio zf619, GenBank: EU126946; Danio zf69, GenBank: EU126948; Danio brd2iso8, GenBank: XM_704349; Danio simbrd2, GenBank: XM_692734.2; Danio chr16.1, 16.2, 16.3, from Ensembl: ENSDARG00000046087; Danio brd2b, GenBank: BC055508; Danio brd3, GenBank: XM_693856; Danio gpm6a, GenBank: BC095027; Danio brd3b, GenBank: NM_212702; Danio brd4, GenBank: XM_694025; Danio brdt, GenBank: XM_701882; Onchorynchus brd2L, GenBank: ABB52829; Takifugu brd2, Ensembl: SINFRUG00000151665; Takifugu brd3, EMBL: AJ311635; Takifugu brd4, Ensembl: SINFRUG00000141925; Takifugu brd4like, Ensembl: SINFRUG00000145405; Takifugu brdt, Ensembl: SINFRUG00000148459; Takifugu brd, Ensembl: SINFRUG00000151665; Tetraodon brd2, EMBL: CAG11678; Tetraodon brd3, EMBL: CAF94980; Tetraodon brd4, EMBL: CAF92198; Tetraodon brd4like, EMBL: CAF90901; Tetraodon brdt, EMBL: CAF96012; Tetraodon brd, EMBL: CAF91369; Oryzias Ring3, GenBank: AB183488; Oryzias brd3, Ensembl: ENSORLG00000016659; Oryzias brd4, Ensembl: ENSORLG00000006490; Oryzias brd4like, Ensembl: ENSORLG00000010329; Oryzias brdt, Ensembl: ENSORLG00000013915; Oryzias brd, Ensembl: ENSORLG00000012190; Oryzias Unknown brd, Ensembl: ENSORLG00000016149; Xenopus brd2, GenBank BC043784; Xenopus brd3, EMBL: CAJ81450; Xenopus brd4, GenBank: BC076786; Xenopus loc443648, GenBank: BC097528; Xenopus brdt, EMBL: CAJ81450; Xenopus loc398944, EMBL: GenBank: BC060452; Gallus brd2, GenBank: NM_001030674; Gallus brd3, GenBank: XM_425330.1; Gallus brd4, Ensembl: ENSGALG00000013295; Gallus brdt, Ensembl: ENSGALG00000006031; Mus Brd2, GenBank: NM_010238; Mus Brd3, GenBank: NM_023336; Mus Brd4iso1, GenBank: NM_020508; Mus Brd4iso2, GenBank: NM_198094; Mus Brdt, GenBank: NM_054054; Homo BRD2, GenBank: NM_005104; Homo BRD3, GenBank: NM_007371; Homo BRD4short, GenBank: NM_014299; Homo BRD4long, GenBank: NM_058243;Homo BRDT, GenBank: NM_207189; Myxine Ring3, GenBank: AF191032; Brachiostoma brd, GenBank: AF391288; Anopheles brd, GenBank: XM_312107; Drosophila fsh, GenBank: NM_206647.
Figure 3
Comparisons of core BET domains in selected Brd2 species homologs. Amino acid sequences of core BET domains from Brd2 orthologs in Homo, Oryzias, Tetraodon, Takifugu, and Danio; and from Brd2 paralogous sequences in Danio (chr16.1 or chr16.2 for simBrd2; and Brd2b), Onchorynchus (Oncho2L for Brd2L) and Oryzias (OryzUn for Brd), are shown below consensus sequences (top line of each section, in bold), for bromodomain 1 (Bromo 1; 60 amino acid core), bromodomain 2 (Bromo 2; 60 amino acid core) and ET domain (ET; first 120 amino acids). Amino acid numbers at right show position relative to entire protein. Residues within domains that differ from consensus are shown in bold. Asteriks (*) denote bromodomain residues important for acetyl lysine-binding [3]. Dots (.) denote residues important for BD1/BD1 dimerization [32]. (--) shows varying residues in paralogs in ZA loop region. Danio is represented by zf626/Brd2 (Danio), and paralogous proteins encoded by ENSDART00000049051 (chr16.1, bromo 1 and 2), GENSCAN00000006161 (chr16.2, ET domain), and BC055508.1 cDNA (Brd2b; ends prematurely before ET domain). Sequences used here are translations of those used to construct phylogenetic trees (see Fig. 3 legend).
Figure 4
brd2a and brd2b encode gonad-enriched RNAs and multiple developmentally regulated transcripts. Northern blot of total RNA (5 μg/lane) from dissected ovary (ov), adult fish (Ad), and embryos of various stages (0–2, 4–6, 8–10, 14–18, 20–24, and 44–48 hour post fertilization) probed sequentially with A) zf626 (brd2) and B) zf69 (brd2b), radiolabeled cDNA subfragments. RNAs were visualized with EtBr staining and were undegraded and of equivalent amounts in each lane (data not shown). The leftmost ovary lane and the rightmost adult lane are underexposed replicates of adjacent lanes, to allow better band visualization. Ovary-enriched brd2 RNAs (A, 4.8 and 2.8 kb) and ovary-enriched brd2b RNA (B, 6.0 kb) are also present in early embryos (A and B, 0–2 h), and are indicated by asterisks (*). Various size classes of brd2 and brd2b RNAs present in whole adults are also detectable in dissected testis (data not shown).
Figure 5
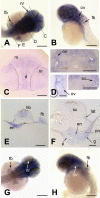
brd2 paralogs are enriched in developing nervous system during segmentation. Whole mount in situ hybridizations to RNA in zebrafish embryos conducted with DIG-labelled zf626 (brd2) (A-L) and zf69 (brd2b) (M-P) cloned sequences. Ubiquitous expression is seen in embryos of 2 cell (A), 16 cell (B), 30% epiboly (C), and bud (D) stages for both brd2 (shown) and brd2b (not shown) genes. Enriched expression in "ladder" patterns along the developing neural keel/rod during segmentation is seen in 14 somite embryos probed for brd2 (E) and for brd2b (M). Cross-sections of 14 somite whole mount embryos probed for brd2 (I, schematic with real image below; 5–6 somite level) show enriched expression in ventrolateral neural rod (nr), floorplate above notochord (nc), and ventral somites (s). Expression is most prominent in head region of 18 somite embyros for both paralogs (F, G; and N, O); only brd2 is also enriched in tail bud (F), while brd2b remains high along the entire ventral trunk (N) at this stage. By 24 hours, brd2 is prominent in the entire head, with some expression in ventral trunk and post-anal tail (H), while brd2b is enriched in ventral brain, cerebellum, and ventral trunk, with low level ubiquitous expression (P). Dorsal flat mounts of 24 hour embryos (J, K) show brd2 expression enriched within caudal tectum (t) at hindbrain-midbrain boundary, within specific cells of the cerebellum (cb) and at the caudal border between cerebellum and rest of hindbrain. The image in (K) is a higher magnification view of a region from another embryo equivalent to the boxed region in J. Expression of brd2 in cells of otic vesicle walls (ov) is apparent both in flat mounts (J) and cross sections at that level (L). Bar = 250 μm for A-H, J, M-P; = 50 μm for I, K; = 100 μm for L.
Figure 6
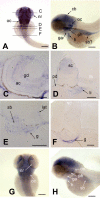
brd2 paralogs are differentially expressed in head and trunk regions of pharyngula stage embryos. Whole mount in situ hybridizations to RNA in 44–48 hour zebrafish embryos with DIG-labelled zf626 (A-F) and zf69 (G, H) cloned sequences. Dorsal (A) and lateral (B) views of whole embyros show brd2 expression abundant in all brain subdivisions and developing pharyngeal arches, and in neural retina (nr), otic vesicle (ov), pectoral fin buds (fb) and ventral trunk. Cross sections at levels indicated in A, reveal brd2 expression in: C) mesencephalon (m), diencephalon (d) and neural retina (nr); D) in cells of otic vesicle walls (ov, black arrows); E) in caudal hindbrain (hb), endoderm of prospective digestive tract (en), and pectoral fin bud (fb); F) in developing spinal cord (sc), posterior lateral line primordium (lat), pronephric duct (pn), and endodermal tissue (en), including pancreas progenitor (p) and developing gut (g). Signal in hollow of otic vesicle (white arrows in D) is an artefact due to trapped probe. Although brd2b (G dorsal, H lateral) is also expressed in head region and pectoral fin buds (fb), it is reduced in dorsal midbrain, caudal hindbrain, and ventral trunk, and absent from neural retina (nr, white arrow) and otic vesicle (ov, white arrow). Bar = 250 μm for A,B,G,H; = 100 μm for C, D (two right images); = 25 μm for D (left image).
Figure 7
brd2 paralogs are differentially expressed in head and digestive system of pecfin stage embryos. Whole mount in situ hybridizations to RNA in 60–65 hour zebrafish embryos with DIG-labelled zf626 (A-F) and zf69 (G, H) cloned sequences. Dorsal (A) and lateral (B) views of whole embryos show brd2 expression in all brain subdivisions, especially cerebellum (cb), in neural retina (nr), otic capsule (oc), atrium of heart (h), gill arches (ga), pharynx (ph), swim bladder (sb) and ventral trunk. Cross sections at levels indicated in A reveal brd2 expression in: C) amacrine cells (ac) and ganglion cell layer (gcl) of neural retina; D) spinal cord (sc), pronephric duct (pd) and endodermal derivatives such as liver (li); E) posterior lateral line primordium (lat), and endodermal derivatives such as swim bladder and gut (g). brd2 expression continues in gut (g), but declines in spinal cord (sc, white) as more posterior sections are assayed (F), and is severely reduced in fin bud (D; fb, white) Expression of brd2b (G dorsal, H lateral) is found mainly in head region, but is reduced overall, especially in hindbrain, and nearly absent from otic capsule (oc, white), gill arches (ga, white), heart (h, white), ventral trunk, and endodermal derivatives such as pharanx (ph, white), and swim bladder (sb, white). Bar = 250 μm for A,B,D,G,H; = 100 μm for C,E,F.
Figure 8
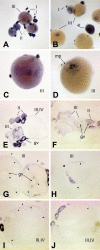
brd2 paralogs are maternal transcripts with different patterns of expression in oocytes. Whole mount in situ hybridizations to RNA in zebrafish ovaries with DIG-labelled zf626 (A, C, E, G, I) and zf69 (B, D, F, H, J) cloned sequences. Cross sections through ovaries are shown in E-J. brd2 maternal RNAs are dispersed and abundant in stage I and II oocytes (A, E), and are sometimes found inside the germinal vesicle (gv), as in G (see small oocytes at far right). By stage III,brd2 transcripts accumulate around the outside of the germinal vesicle and rather unevenly around the periphery of the cortex just inside the vitelline envelope (C, E; G, arrowheads). brd2 RNAs then localize around the periphery of the cortex by late stage III, early stage IV (I, arrowheads). brd2 RNAs are also present in follicle cells of stage III/IV oocytes (C; G, I, arrowheads outside oocyte). Although brd2b maternal transcripts are also dispersed throughout stage I and II oocytes, and often found inside the germinal vesicle (B, F, H, J), in contrast to brd2, they become strictly localized to the region surrounding the micropyle (D, mp; H, J, arrowheads), the future animal pole, sometime during stage III, and are not expressed in follicle cells. A,B: 40X; C-F: 80X; G-J, 100X.