Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes - PubMed (original) (raw)
. 2008 May 22;453(7194):534-8.
doi: 10.1038/nature06904. Epub 2008 Apr 10.
Affiliations
- PMID: 18404147
- PMCID: PMC2981145
- DOI: 10.1038/nature06904
Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes
Oliver H Tam et al. Nature. 2008.
Abstract
Pseudogenes populate the mammalian genome as remnants of artefactual incorporation of coding messenger RNAs into transposon pathways. Here we show that a subset of pseudogenes generates endogenous small interfering RNAs (endo-siRNAs) in mouse oocytes. These endo-siRNAs are often processed from double-stranded RNAs formed by hybridization of spliced transcripts from protein-coding genes to antisense transcripts from homologous pseudogenes. An inverted repeat pseudogene can also generate abundant small RNAs directly. A second class of endo-siRNAs may enforce repression of mobile genetic elements, acting together with Piwi-interacting RNAs. Loss of Dicer, a protein integral to small RNA production, increases expression of endo-siRNA targets, demonstrating their regulatory activity. Our findings indicate a function for pseudogenes in regulating gene expression by means of the RNA interference pathway and may, in part, explain the evolutionary pressure to conserve argonaute-mediated catalysis in mammals.
Figures
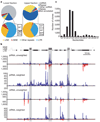
Figure 1. Both piRNA and siRNA systems control transposons in mouse oocytes
a, Small RNA libraries from 19–24-nucleotide (lower fraction, left) and 24–30-nucleotide RNAs (upper fraction, right) were deeply sequenced. Reads were assigned an annotation as previously described. The fraction of reads in each category is depicted. The repeat-annotated small RNAs were designated as LINE, SINE (short interspersed element), LTR and other, with the LTR category further divided between MT (mouse transcript), IAP and other. b, A representative piRNA locus on chromosome 10 is shown with the content of LINE, SINE and LTR fragments. Shading indicates the degree of match to the consensus element. Frequency plots for piRNAs and siRNAs from this locus are shown below. ‘Unweighted’ plots each match to the cluster; ‘weighted’ normalizes each match, dividing by its genomic frequency. Blue and red lines indicate small RNAs mapping to the upper and lower genomic strand, respectively. c, RNAs from 19–30 nucleotides were deeply sequenced. Reads matching the piRNA cluster shown in b were used to construct a frequency plot by length.
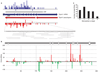
Figure 2. Gene–pseudogene interactions produce endogenous siRNAs
a, Endo-siRNAs unambiguously mapped to the functional Ppp4r1 mRNA are plotted in blue, above the mRNA (individual exons indicated as thick arrows). The extent of the open reading frame (ORF) is indicated. Endo-siRNAs from the Ppp4r1 pseudogene are shown in red below the pseudogene. Arrows indicate two segments of Ppp4r1 homology. siRNAs plotted above each line are sense oriented, with respect to the functional mRNA, and those below are antisense. Shown below is an enlargement of one section of the mRNA with individual antisense siRNAs aligned. A dash indicates a match; an asterisk indicates a mismatch. The heights of the bars indicate the number of siRNAs starting at each position. b, Endogenous siRNAs homologous to Ran-GAP are shown below a schematic of the genomic inverted repeat structure from which they arise (the ~800-base loop is not depicted). Those shown above and below the x axis come from the upper or lower arm of the hairpin, respectively. No siRNAs were sequenced from the ~800-base loop that separates the two stem arms. c, siRNAs antisense to Ppp4r1 counted and plotted by the number of mismatches to the functional mRNA.
Figure 3. Endo-siRNAs have a role in gene regulation
a, We had previously compared expression levels in wild-type and Dicer-null oocytes by microarray. Genes with a large number of siRNAs were examined. For those with significant changes in expression (P < 0.1), the fold change in Dicer-null (KO) versus wild type (WT) was plotted. The graph was arranged according to the number of antisense siRNAs per gene in our data set, increasing from left to right (with benchmarks shown). The identity of the gene represented by each bar is given below. b, Fourteen genes (with and without siRNAs in our data set, indicated by siRNA+ and siRNA−, respectively), comprising a set in which some showed significant changes and some did not, were tested for changes in mRNA levels in Dicer-null versus wild-type oocytes by quantitative PCR. Gene names are indicated below each bar; error bars represent the mean ± s.e.m. (three replicates).
Similar articles
- Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes.
Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, Chiba H, Kohara Y, Kono T, Nakano T, Surani MA, Sakaki Y, Sasaki H. Watanabe T, et al. Nature. 2008 May 22;453(7194):539-43. doi: 10.1038/nature06908. Epub 2008 Apr 10. Nature. 2008. PMID: 18404146 - Small RNAs originated from pseudogenes: cis- or trans-acting?
Guo X, Zhang Z, Gerstein MB, Zheng D. Guo X, et al. PLoS Comput Biol. 2009 Jul;5(7):e1000449. doi: 10.1371/journal.pcbi.1000449. Epub 2009 Jul 31. PLoS Comput Biol. 2009. PMID: 19649160 Free PMC article. - The ERI-6/7 helicase acts at the first stage of an siRNA amplification pathway that targets recent gene duplications.
Fischer SE, Montgomery TA, Zhang C, Fahlgren N, Breen PC, Hwang A, Sullivan CM, Carrington JC, Ruvkun G. Fischer SE, et al. PLoS Genet. 2011 Nov;7(11):e1002369. doi: 10.1371/journal.pgen.1002369. Epub 2011 Nov 10. PLoS Genet. 2011. PMID: 22102828 Free PMC article. - Roles of piRNAs in transposon and pseudogene regulation of germline mRNAs and lncRNAs.
Wang C, Lin H. Wang C, et al. Genome Biol. 2021 Jan 8;22(1):27. doi: 10.1186/s13059-020-02221-x. Genome Biol. 2021. PMID: 33419460 Free PMC article. Review. - Endogenous siRNAs: regulators of internal affairs.
Piatek MJ, Werner A. Piatek MJ, et al. Biochem Soc Trans. 2014 Aug;42(4):1174-9. doi: 10.1042/BST20140068. Biochem Soc Trans. 2014. PMID: 25110021 Free PMC article. Review.
Cited by
- Human Transcriptome and Chromatin Modifications: An ENCODE Perspective.
Shen L, Choi I, Nestler EJ, Won KJ. Shen L, et al. Genomics Inform. 2013 Jun;11(2):60-7. doi: 10.5808/GI.2013.11.2.60. Epub 2013 Jun 30. Genomics Inform. 2013. PMID: 23843771 Free PMC article. - Double-stranded RNA made in C. elegans neurons can enter the germline and cause transgenerational gene silencing.
Devanapally S, Ravikumar S, Jose AM. Devanapally S, et al. Proc Natl Acad Sci U S A. 2015 Feb 17;112(7):2133-8. doi: 10.1073/pnas.1423333112. Epub 2015 Feb 2. Proc Natl Acad Sci U S A. 2015. PMID: 25646479 Free PMC article. - Epigenetic silencing of Oct4 by a complex containing SUV39H1 and Oct4 pseudogene lncRNA.
Scarola M, Comisso E, Pascolo R, Chiaradia R, Marion RM, Schneider C, Blasco MA, Schoeftner S, Benetti R. Scarola M, et al. Nat Commun. 2015 Jul 9;6:7631. doi: 10.1038/ncomms8631. Nat Commun. 2015. PMID: 26158551 Free PMC article. - Mice lacking microRNAs in Pax8-expressing cells develop hypothyroidism and end-stage renal failure.
Bartram MP, Amendola E, Benzing T, Schermer B, de Vita G, Müller RU. Bartram MP, et al. BMC Mol Biol. 2016 Apr 18;17:11. doi: 10.1186/s12867-016-0064-x. BMC Mol Biol. 2016. PMID: 27090781 Free PMC article. - Tupaia small RNAs provide insights into function and evolution of RNAi-based transposon defense in mammals.
Rosenkranz D, Rudloff S, Bastuck K, Ketting RF, Zischler H. Rosenkranz D, et al. RNA. 2015 May;21(5):911-22. doi: 10.1261/rna.048603.114. Epub 2015 Mar 23. RNA. 2015. PMID: 25802409 Free PMC article.
References
- D’Errico I, Gadaleta G, Saccone C. Pseudogenes in metazoa: origin and features. Brief. Funct. Genomics Proteomics. 2004;3:157–167. - PubMed
- Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007;318:761–764. - PubMed
- Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007;316:744–747. - PubMed
- Brennecke J, et al. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. - PubMed
- Carmell MA, et al. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev. Cell. 2007;12:503–514. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases