The human ABC transporter pseudogene family: Evidence for transcription and gene-pseudogene interference - PubMed (original) (raw)
Comparative Study
The human ABC transporter pseudogene family: Evidence for transcription and gene-pseudogene interference
Armin P Piehler et al. BMC Genomics. 2008.
Abstract
Background: Pseudogenes are an integral component of the human genome. Little attention, however, has so far been paid to the phenomenon that some pseudogenes are transcriptionally active. Recently, we demonstrated that the human ortholog of the rodent testis-specific ATP-binding cassette (ABC) transporter Abca17 is a ubiquitously transcribed pseudogene (ABCA17P). The aim of the present study was to establish a complete inventory of all ABC transporter pseudogenes in the human genome and to identify transcriptionally active ABC transporter pseudogenes. Moreover, we tested the hypothesis that a regulatory interdependency exists between ABC transporter pseudogenes and their parental protein coding equivalents.
Results: Systematic bioinformatic analysis revealed the existence of 22 ABC transporter pseudogenes within the human genome. We identified two clusters on chromosomes 15 and 16, respectively, which harbor almost half of all pseudogenes (n = 10). Available information from EST and mRNA databases and RT-PCR expression profiling indicate that a large portion of the ABC transporter pseudogenes (45%, n = 10) are transcriptionally active and some of them are expressed as alternative splice variants. We demonstrate that both pseudogenes of the pseudoxanthoma elasticum gene ABCC6, ABCC6P1 and ABCC6P2, are transcribed. ABCC6P1 and ABCC6 possess near-identical promoter sequences and their tissue-specific expression profiles are strikingly similar raising the possibility that they form a gene-pseudogene dual transcription unit. Intriguingly, targeted knockdown of the transcribed pseudogene ABCC6P1 resulted in a significant reduction of ABCC6 mRNA expression levels.
Conclusion: The human genome contains a surprisingly small number of ABC transporter pseudogenes relative to other known gene families. They are unevenly distributed across the chromosomes. Importantly, a significant portion of the ABC transporter pseudogenes is transcriptionally active. The downregulation of ABCC6 mRNA levels by targeted suppression of the expression of its pseudogene ABCC6P1 provides evidence, for the first time, for a regulatory interdependence of a transcribed pseudogene and its protein coding counterpart in the human genome.
Figures
Figure 1
Chromosomal distribution of human ABC transporter pseudogenes and protein-coding genes. Pseudogenes of the human ABC transporter family are widely distributed throughout the human genome. Shown are the chromosomal localizations (colored arrows) of ABC transporter pseudogenes (right-hand side of each chromosome) and their parental protein-coding genes (left) in the human genome. Pairs of parental genes and pseudogenes are represented by the same arrow color code. Transcribed pseudogenes are green shaded. A close-up view of the ABC transporter pseudogene cluster (30 Mb) on chromosome 16p is shown at the bottom. Arrows indicate the 5'-> 3' orientation of the pseudogenes on chr. 16p and distances between adjacent pseudogenes are detailed (Mb). ABCA14P, ABCA15P1 and ABCA15P2, respectively, represent pseudogenes for which functional counterparts have only been reported in rodents (Abca14, Abca15). The ideogram was generated using the ColoredChromosomes software [41].
Figure 2
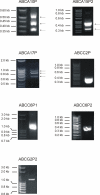
Expression of human ABC transporter pseudogene RNAs. RT-PCR expression profiling demonstrating that the ABC transporter pseudogenes ABCA10P, ABCA15P2, ABCA17P, ABCC2P, ABCC6P1, ABCC6P2 and ABCG2P2, respectively, are transcribed as predicted by our in silico results.RT-PCR was performed using human cDNA from pooled tissues. Note that ABCA10P, ABCA15P2, and ABCA17P are expressed as alternatively spliced transcript variants (as confirmed by sequence analysis) (arrows).
Figure 3
Genomic organization of ABCC6P1 and ABCC6P2, two transcribed pseudogenes of ABCC6. The pseudogenes ABCC6P1 and ABCC6P2 flank their parental gene ABCC6 at a distance of 2.3 Mb and 1.3 Mb, respectively, on chromosome 16p13. Arrows indicate the direction of transcription. The structures of the ABCC6P1 and ABCC6P2 genes and their transcripts are illustrated in higher magnification. Exons are represented by black boxes and numbered in 5' to 3' order. Similarities (%) of pseudogene exons with their corresponding ABCC6 exons are shown in parenthesis. The terminal non-homologous exon of ABCC6P1 is shaded blue. The red box highlights the last exon of ABCC6P2 which contains a part of intron 2. Metric scale bars are indicated for orientation.
Figure 4
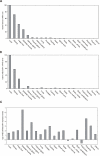
ABCC6 and its transcribed pseudogene ABCC6P1 are co-expressed in a variety of human tissues. Relative transcription levels of ABCC6 (A) and ABCC6P1 (B) were determined in 20 different human tissues by real-time RT-PCR relative to those of the housekeeping gene β-actin. Highest expression of both ABCC6 and ABCC6P1 was observed in adult liver (reference = 100%), kidney and fetal liver. (C) ABCC6/ABCC6P1 transcript ratios. Note that ABCC6 and ABCC6P1 are co-expressed in all tissues investigated with the ABCC6 expression levels being on average 2–4 times higher than those of ABCC6P1. Ratios are shown in a logarithmic scale (base 2).
Figure 5
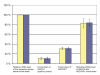
Silencing of the transcribed pseudogene ABCC6P1 downregulates mRNA levels of ABCC6. Targeted siRNA-mediated inhibition of ABCC6P1 expression in HepG2 cells results in significant downregulation (69%, 95%-CI: 65–73%) of the pseudogene transcript levels (lane 3). mRNA levels are normalized to β-actin (yellow bars) and cyclophilin B (blue bars). ABCC6P1 silencing is associated with a reduction of ABCC6 mRNA expression (84%, p = 0.003, 95%-CI: 76–91%, yellow bar; 82%, p = 0.01, 95%-CI: 72–93%, blue bar, lane 4). A positive control (knock-down of GAPDH) is shown (lane 2). Data were obtained from six independent experiments. The relative expression levels before knock-down experiments were set to 100% (lane 1).
Similar articles
- The human ortholog of the rodent testis-specific ABC transporter Abca17 is a ubiquitously expressed pseudogene (ABCA17P) and shares a common 5' end with ABCA3.
Piehler AP, Wenzel JJ, Olstad OK, Haug KB, Kierulf P, Kaminski WE. Piehler AP, et al. BMC Mol Biol. 2006 Sep 12;7:28. doi: 10.1186/1471-2199-7-28. BMC Mol Biol. 2006. PMID: 16968533 Free PMC article. - Copy number variations of the ATP-binding cassette transporter ABCC6 gene and its pseudogenes.
Kringen MK, Stormo C, Grimholt RM, Berg JP, Piehler AP. Kringen MK, et al. BMC Res Notes. 2012 Aug 9;5:425. doi: 10.1186/1756-0500-5-425. BMC Res Notes. 2012. PMID: 22873774 Free PMC article. - ABCC6P1 pseudogene induces ABCC6 upregulation and multidrug resistance in breast cancer.
Hashemi M, Golalipour M. Hashemi M, et al. Mol Biol Rep. 2022 Oct;49(10):9633-9639. doi: 10.1007/s11033-022-07872-6. Epub 2022 Aug 28. Mol Biol Rep. 2022. PMID: 36030475 - Evolution of the ATP-binding cassette (ABC) transporter superfamily in vertebrates.
Dean M, Annilo T. Dean M, et al. Annu Rev Genomics Hum Genet. 2005;6:123-42. doi: 10.1146/annurev.genom.6.080604.162122. Annu Rev Genomics Hum Genet. 2005. PMID: 16124856 Review. - The emerging role of pseudogene expressed non-coding RNAs in cellular functions.
Groen JN, Capraro D, Morris KV. Groen JN, et al. Int J Biochem Cell Biol. 2014 Sep;54:350-5. doi: 10.1016/j.biocel.2014.05.008. Epub 2014 May 16. Int J Biochem Cell Biol. 2014. PMID: 24842102 Free PMC article. Review.
Cited by
- LncRNA ABCC6P1 promotes proliferation and migration of papillary thyroid cancer cells via Wnt/β-catenin signaling pathway.
Guan Y, Li Y, Yang QB, Yu J, Qiao H. Guan Y, et al. Ann Transl Med. 2021 Apr;9(8):664. doi: 10.21037/atm-21-505. Ann Transl Med. 2021. PMID: 33987362 Free PMC article. - Role of the long non‑coding RNA‑Annexin A2 pseudogene 3/Annexin A2 signaling pathway in biliary atresia‑associated hepatic injury.
Nuerzhati Y, Dong R, Song Z, Zheng S. Nuerzhati Y, et al. Int J Mol Med. 2019 Feb;43(2):739-748. doi: 10.3892/ijmm.2018.4023. Epub 2018 Dec 11. Int J Mol Med. 2019. PMID: 30569159 Free PMC article. - Assessing the genomic evidence for conserved transcribed pseudogenes under selection.
Khachane AN, Harrison PM. Khachane AN, et al. BMC Genomics. 2009 Sep 15;10:435. doi: 10.1186/1471-2164-10-435. BMC Genomics. 2009. PMID: 19754956 Free PMC article. - Human ATP-binding cassette (ABC) transporter family.
Vasiliou V, Vasiliou K, Nebert DW. Vasiliou V, et al. Hum Genomics. 2009 Apr;3(3):281-90. doi: 10.1186/1479-7364-3-3-281. Hum Genomics. 2009. PMID: 19403462 Free PMC article. - Carboxylesterase 1 gene duplication and mRNA expression in adipose tissue are linked to obesity and metabolic function.
Friedrichsen M, Poulsen P, Wojtaszewski J, Hansen PR, Vaag A, Rasmussen HB. Friedrichsen M, et al. PLoS One. 2013;8(2):e56861. doi: 10.1371/journal.pone.0056861. Epub 2013 Feb 28. PLoS One. 2013. PMID: 23468884 Free PMC article.
References
- Kaminski WE, Piehler A, Wenzel JJ. ABC A-subfamily transporters: structure, function and disease. Biochim Biophys Acta. 2006;1762:510–524. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials