Assembly and channel opening in a bacterial drug efflux machine - PubMed (original) (raw)
Assembly and channel opening in a bacterial drug efflux machine
Vassiliy N Bavro et al. Mol Cell. 2008.
Abstract
Drugs and certain proteins are transported across the membranes of Gram-negative bacteria by energy-activated pumps. The outer membrane component of these pumps is a channel that opens from a sealed resting state during the transport process. We describe two crystal structures of the Escherichia coli outer membrane protein TolC in its partially open state. Opening is accompanied by the exposure of three shallow intraprotomer grooves in the TolC trimer, where our mutagenesis data identify a contact point with the periplasmic component of a drug efflux pump, AcrA. We suggest that the assembly of multidrug efflux pumps is accompanied by induced fit of TolC driven mainly by accommodation of the periplasmic component.
Figures
Figure 1
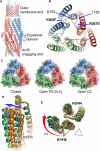
The TolC Outer Membrane Protein and Its Partial Opening (A) A side view of the TolC homotrimer (1EK9). (B) A schematic of the mutations that were studied here to stabilize channel opening. A network of charged interactions maintaining the resting closed state of TolC (1EK9). These include Y362, R367 from H7/H8, which are coordinated by T152, and D153 from H3/H4. Y362 has been mutated to F and R367 to E to disrupt the network of salt bridges (shown in bold). The protomers are colored red, blue, and green. (C) Crystal structures of the open and closed states. The view is along the trifold axis at the periplasmic, AcrB-engaging end of the TolC trimer. (D) Helical movements from the open to the closed state for the two crystal forms. Transition of the mobile helices H7/H8 from closed state (orange) to open as exemplified by the different subunits of C2 (cyan and green) and P212121 (gray and yellow). The view is of overlays of helical fragments H7/H8 (foreground) and H3/H4 (background), revealing the minimal relative movement of H3/H4 static helices as compared with H7/H8. (E) Top view of the same overlays shown in (D). The displacement of the H7 helix is up to 11 Å in the C2 structure. Note also the lagging of the H8 helices and the relative swing of the H7 in respect to H8. The triangle indicates the molecular trifold axis.
Figure 2
Potential Docking Faces of the Partially Opened State of TolC (A and B) The second aperture of the channel is less perturbed in the crystal structures of the partially opened state. A cross-section through the TolC channel in the closed-state (A) and open-state C2 crystal form (B) at the level of the second selectivity filter formed by the ring of D374 residues. While the outer opening of the channel (as measured by the position of G365) from the 8.5 Å in closed state to about 20 Å in the C2 form (Figure 1C, Figure S2), the interior second selectivity filter composed of a ring of D374 deeper in the channel is much less perturbed. Although the distance between the D374 is extended from about 6.1 Å in 1EK9 to up to 8.4 Å in the C2 form, it is unlikely to be sufficient for even small molecules to pass unimpeded, thus suggesting that a further opening of the channel is required for transport. This is likely to be activated by the engagement of the periplasmic partner protein, AcrA. Note the deepening of the predicted AcrA binding groove in the partially open C2 structure. (C) The intermesh of the loops in the packing of the TolC open state, showing details of the trimer-trimer contact interface across a crystallographic symmetry operation in the C2 crystal form. This interaction may mimic the docking of the TolC into the matching surface of the AcrB (shown in [D]). (D) Docking model of AcrB and a model of open-state TolC based on the C2 crystal structure. Colored by chain. The model of the AcrB-TolC complex was prepared using the asymmetric structures (2GIF and C2 crystal form of TolC). Although β2 hairpin is in proximity of H7/H8, it is still capable of interacting with the H3/H4 residues, in agreement with crosslinking data (Tamura et al., 2005). Residues indicated by arrows are D153, one of the residues included in TolC wild-type that maintains the closed gate; D795 from AcrB, a residue from AcrB β2 hairpin, which could potentially disrupt D153 interactions; and Y362 (another gating residue from TolC) and D256 (from β1 hairpin of AcrB), which in our refined docking model are close to the interface.
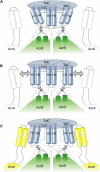
Figure 3
A Cartoon Schematic of the Assembly of the Efflux Pump and Accommodation of AcrA by TolC (A) AcrB docking of TolC results in intermeshing of the exposed crown and helical protrusions. “Mobile” H7/H8 helices surround one “static” helix pair H3/H4 in the middle. Two salt bridges (small dotted lines) support the closed state (as represented by the bent helices). Horizontal dotted line higher up in the TolC channel represents second selectivity filter. Interaction of the TolC H7/H8 helices with the β hairpins of the AcrB crown is represented with two-sided arrows. The AcrA (outlined) may be predocked on the AcrB. (B) Unlocking of bridges. Interaction of β hairpins 1 and 2 in the crown of AcrB with the helical turns of H7/H8 helices from TolC breaks the intramolecular salt bridges, releasing the H7/H8 helices and partially opening the iris of the outer periplasmic entrance of the TolC channel and deepening of the surface grooves allowing the engagement of the AcrA (indicated by two-sided arrows). This AcrB interaction, however, does not affect the second selectivity filter. (C) AcrA binds into the surface grooves of TolC, disrupting the second selectivity filter and opening the channel to its full extent. The machinery is now fully assembled and active for transport.
Similar articles
- Structure of the AcrAB-TolC multidrug efflux pump.
Du D, Wang Z, James NR, Voss JE, Klimont E, Ohene-Agyei T, Venter H, Chiu W, Luisi BF. Du D, et al. Nature. 2014 May 22;509(7501):512-5. doi: 10.1038/nature13205. Epub 2014 Apr 20. Nature. 2014. PMID: 24747401 Free PMC article. - Funnel-like hexameric assembly of the periplasmic adapter protein in the tripartite multidrug efflux pump in gram-negative bacteria.
Xu Y, Lee M, Moeller A, Song S, Yoon BY, Kim HM, Jun SY, Lee K, Ha NC. Xu Y, et al. J Biol Chem. 2011 May 20;286(20):17910-20. doi: 10.1074/jbc.M111.238535. Epub 2011 Mar 29. J Biol Chem. 2011. PMID: 21454662 Free PMC article. - The C-terminal domain of AcrA is essential for the assembly and function of the multidrug efflux pump AcrAB-TolC.
Ge Q, Yamada Y, Zgurskaya H. Ge Q, et al. J Bacteriol. 2009 Jul;191(13):4365-71. doi: 10.1128/JB.00204-09. Epub 2009 May 1. J Bacteriol. 2009. PMID: 19411330 Free PMC article. - Three's company: component structures bring a closer view of tripartite drug efflux pumps.
Eswaran J, Koronakis E, Higgins MK, Hughes C, Koronakis V. Eswaran J, et al. Curr Opin Struct Biol. 2004 Dec;14(6):741-7. doi: 10.1016/j.sbi.2004.10.003. Curr Opin Struct Biol. 2004. PMID: 15582398 Review. - RND efflux pumps: structural information translated into function and inhibition mechanisms.
Ruggerone P, Murakami S, Pos KM, Vargiu AV. Ruggerone P, et al. Curr Top Med Chem. 2013;13(24):3079-100. doi: 10.2174/15680266113136660220. Curr Top Med Chem. 2013. PMID: 24200360 Review.
Cited by
- Structural shifts in TolC facilitate Efflux-Mediated β-lactam resistance.
Kantarcioglu I, Gaszek IK, Guclu TF, Yildiz MS, Atilgan AR, Toprak E, Atilgan C. Kantarcioglu I, et al. Commun Biol. 2024 Aug 26;7(1):1051. doi: 10.1038/s42003-024-06750-0. Commun Biol. 2024. PMID: 39187619 Free PMC article. - Structures and Efflux Mechanisms of the AcrAB-TolC Pump.
Yu Z, Shi X, Wang Z. Yu Z, et al. Subcell Biochem. 2024;104:1-16. doi: 10.1007/978-3-031-58843-3_1. Subcell Biochem. 2024. PMID: 38963480 Review. - The Mla system of diderm Firmicute Veillonella parvula reveals an ancestral transenvelope bridge for phospholipid trafficking.
Grasekamp KP, Beaud Benyahia B, Taib N, Audrain B, Bardiaux B, Rossez Y, Izadi-Pruneyre N, Lejeune M, Trivelli X, Chouit Z, Guerardel Y, Ghigo JM, Gribaldo S, Beloin C. Grasekamp KP, et al. Nat Commun. 2023 Nov 23;14(1):7642. doi: 10.1038/s41467-023-43411-y. Nat Commun. 2023. PMID: 37993432 Free PMC article. - HyperVR: a hybrid deep ensemble learning approach for simultaneously predicting virulence factors and antibiotic resistance genes.
Ji B, Pi W, Liu W, Liu Y, Cui Y, Zhang X, Peng S. Ji B, et al. NAR Genom Bioinform. 2023 Feb 11;5(1):lqad012. doi: 10.1093/nargab/lqad012. eCollection 2023 Mar. NAR Genom Bioinform. 2023. PMID: 36789031 Free PMC article. - A coevolution experiment between Flavobacterium johnsoniae and Burkholderia thailandensis reveals parallel mutations that reduce antibiotic susceptibility.
Chodkowski JL, Shade A. Chodkowski JL, et al. Microbiology (Reading). 2023 Feb;169(2):001267. doi: 10.1099/mic.0.001267. Microbiology (Reading). 2023. PMID: 36724091 Free PMC article.
References
- Akama H., Matsuura T., Kashiwagi S., Yoneyama H., Narita S., Tsukihara T., Nakagawa A., Nakae T. Crystal structure of the membrane fusion protein, MexA, of the multidrug transporter in Pseudomonas aeruginosa. J. Biol. Chem. 2004;279:25939–25942. - PubMed
- Bokma E., Koronakis E., Lobedanz S., Hughes C., Koronakis V. Directed evolution of a bacterial efflux pump: adaptation of the E. coli TolC exit duct to the Pseudomonas MexAB translocase. FEBS Lett. 2006;580:5339–5343. - PubMed
- CCP4 (Collaborative Computational Project, Number 4) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases