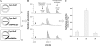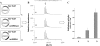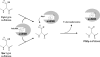Anaerobic sulfatase-maturating enzymes, first dual substrate radical S-adenosylmethionine enzymes - PubMed (original) (raw)
Anaerobic sulfatase-maturating enzymes, first dual substrate radical S-adenosylmethionine enzymes
Alhosna Benjdia et al. J Biol Chem. 2008.
Abstract
Sulfatases are a major group of enzymes involved in many critical physiological processes as reflected by their broad distribution in all three domains of life. This class of hydrolases is unique in requiring an essential post-translational modification of a critical active-site cysteine or serine residue to C(alpha)-formylglycine. This modification is catalyzed by at least three nonhomologous enzymatic systems in bacteria. Each enzymatic system is currently considered to be dedicated to the modification of either cysteine or serine residues encoded in the sulfatase-active site and has been accordingly categorized as Cys-type and Ser-type sulfatase-maturating enzymes. We report here the first detailed characterization of two bacterial anaerobic sulfatase-maturating enzymes (anSMEs) that are physiologically responsible for either Cys-type or Ser-type sulfatase maturation. The activity of both enzymes was investigated in vivo and in vitro using synthetic substrates and the successful purification of both enzymes facilitated the first biochemical and spectroscopic characterization of this class of enzyme. We demonstrate that reconstituted anSMEs are radical S-adenosyl-l-methionine enzymes containing a redox active [4Fe-4S](2+,+) cluster that initiates the radical reaction by binding and reductively cleaving S-adenosyl-l-methionine to yield 5 '-deoxyadenosine and methionine. Surprisingly, our results show that anSMEs are dual substrate enzymes able to oxidize both cysteine and serine residues to C(alpha)-formylglycine. Taken together, the results support a radical modification mechanism that is initiated by hydrogen abstraction from a serine or cysteine residue located in an appropriate target sequence.
Figures
FIGURE 1.
Sequence alignment of the three anSME putative clusters, AtsB (K. pneumoniae), anSMEcpe (CPF_0616, C. perfringens), and anSMEbt (BT_0238, B. thetaiotaomicron). Sequence positions in the proteins are in brackets. In black are the conserved cysteines and in gray are the other conserved residues.
FIGURE 2.
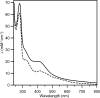
A, gel electrophoresis analysis of anSMEcpe (lane 1) and anSMEbt (lane 2)(MW, molecular weight markers). B, UV-visible absorption spectra of reconstituted anSMEbt (dotted line) and anSMEcpe (solid line).
FIGURE 3.
UV-visible absorption spectra of as isolated (dashed line) and reconstituted (solid line) anSMEcpe.
FIGURE 4.
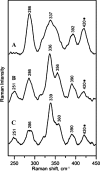
Resonance Raman spectra of anSMEcpe as isolated (A), reconstituted (B), and reconstituted in the presence of a 20-fold excess of SAM (C). The spectra were recorded with 458-nm excitation, using samples that were ∼3 m
m
in anSMEcpe frozen at 20 K, with 140 milliwatt laser power at the sample. Each scan involved photon counting for 1 s at 1.0 cm–1 increments with 7 cm–1 spectral resolution, and each spectrum is the sum of ∼100 scans. Bands resulting from the lattice modes have been subtracted from each spectrum and the band at 420 cm–1 that is marked with as asterisk contains a contribution from glycerol.
FIGURE 5.
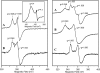
EPR spectra of anSMEcpe after anaerobic reduction with a 10-fold excess of sodium dithionite. Spectra were recorded for samples of 0.2 m
m
anSMEcpe at 10 K with a microwave frequency of 9.603 GHz, a modulation amplitude of 0.63 millitesla, and a microwave power of 10 milliwatt, unless otherwise indicated. Left panel, reconstituted anSMEcpe (A) and as isolated anSMEcpe (B). Inset shows the low field region of the spectrum for reconstituted anSMEcpe recorded at 4.3 K and 50 m
m
. Right panel, reconstituted anSMEcpe in the presence (A) or absence (B) of a 20-fold excess of SAM. The difference spectrum corresponding to: A minus 0.5 ×B is shown in C.
FIGURE 6.
AdoMet reductive cleavage assayed by reverse phase HPLC. Incubation of 20 μ
m
reconstituted anSMEbt (A) or anSMEcpe (B), with 1 m
m
AdoMet alone (□) or with 23-mer peptides (500 μ
m
) containing a cysteine (♦), serine (▴), or alanine (▪) as target residue.
FIGURE 7.
In vitro maturation of 23-mer peptides with reconstituted anSMEcpe. After 6 h of incubation under anaerobic conditions in the presence of AdoMet and a cysteine (panel 1), serine (panel 2), or alanine-containing peptide (panel 3), maturation was analyzed by MALDI-TOF MS either with a α-cyano-4-hydroxycinnamic acid (A) or a DNPH matrix (B).
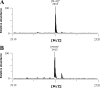
FIGURE 8.
MALDI-TOF MS analysis of the serine (A) and FGly-containing peptide (B) purified by HPLC.
FIGURE 9.
In vivo_maturation of_C. perfringens sulfatase expressed alone (panel 1) or in the presence of anSMEcpe (panel 2) or anSMEbt (panel 3). A, maps of the plasmids used for the anaerobic production of the C. perfringens sulfatase. B, MALDI-TOF MS analysis of purified sulfatase digested with trypsin and CNBr (numbers indicate the amino acid residues). C, specific activity of the purified sulfatases.
FIGURE 10.
In vivo maturation of the serine mutant of the C. perfringens sulfatase expressed alone (panel 1) or in the presence of anSMEcpe (panel 2) or anSMEbt (panel 3). A, maps of the plasmids used for the anaerobic production of the serine mutant of C. perfringens sulfatase. B, MALDI-TOF MS analysis of purified sulfatases digested with trypsin and CNBr (numbers indicate the amino acid residues). C, specific activity of the purified sulfatases.
FIGURE 11.
Proposed mechanism for the maturation reaction catalyzed by anSMEs.
Similar articles
- Anaerobic sulfatase-maturating enzyme--a mechanistic link with glycyl radical-activating enzymes?
Benjdia A, Subramanian S, Leprince J, Vaudry H, Johnson MK, Berteau O. Benjdia A, et al. FEBS J. 2010 Apr;277(8):1906-20. doi: 10.1111/j.1742-4658.2010.07613.x. Epub 2010 Mar 9. FEBS J. 2010. PMID: 20218986 Free PMC article. - A new type of bacterial sulfatase reveals a novel maturation pathway in prokaryotes.
Berteau O, Guillot A, Benjdia A, Rabot S. Berteau O, et al. J Biol Chem. 2006 Aug 11;281(32):22464-70. doi: 10.1074/jbc.M602504200. Epub 2006 Jun 9. J Biol Chem. 2006. PMID: 16766528 - Mechanistic investigations of anaerobic sulfatase-maturating enzyme: direct Cbeta H-atom abstraction catalyzed by a radical AdoMet enzyme.
Benjdia A, Leprince J, Sandström C, Vaudry H, Berteau O. Benjdia A, et al. J Am Chem Soc. 2009 Jun 24;131(24):8348-9. doi: 10.1021/ja901571p. J Am Chem Soc. 2009. PMID: 19489556 - Pyruvate formate-lyase activating enzyme: elucidation of a novel mechanism for glycyl radical formation.
Buis JM, Broderick JB. Buis JM, et al. Arch Biochem Biophys. 2005 Jan 1;433(1):288-96. doi: 10.1016/j.abb.2004.09.028. Arch Biochem Biophys. 2005. PMID: 15581584 Review. - SPASM and twitch domains in S-adenosylmethionine (SAM) radical enzymes.
Grell TA, Goldman PJ, Drennan CL. Grell TA, et al. J Biol Chem. 2015 Feb 13;290(7):3964-71. doi: 10.1074/jbc.R114.581249. Epub 2014 Dec 4. J Biol Chem. 2015. PMID: 25477505 Free PMC article. Review.
Cited by
- Stoichiometry of the redox neutral deamination and oxidative dehydrogenation reactions catalyzed by the radical SAM enzyme DesII.
Ruszczycky MW, Choi SH, Liu HW. Ruszczycky MW, et al. J Am Chem Soc. 2010 Feb 24;132(7):2359-69. doi: 10.1021/ja909451a. J Am Chem Soc. 2010. PMID: 20121093 Free PMC article. - Characterization of glycosaminoglycan (GAG) sulfatases from the human gut symbiont Bacteroides thetaiotaomicron reveals the first GAG-specific bacterial endosulfatase.
Ulmer JE, Vilén EM, Namburi RB, Benjdia A, Beneteau J, Malleron A, Bonnaffé D, Driguez PA, Descroix K, Lassalle G, Le Narvor C, Sandström C, Spillmann D, Berteau O. Ulmer JE, et al. J Biol Chem. 2014 Aug 29;289(35):24289-303. doi: 10.1074/jbc.M114.573303. Epub 2014 Jul 7. J Biol Chem. 2014. PMID: 25002587 Free PMC article. - Further characterization of Cys-type and Ser-type anaerobic sulfatase maturating enzymes suggests a commonality in the mechanism of catalysis.
Grove TL, Ahlum JH, Qin RM, Lanz ND, Radle MI, Krebs C, Booker SJ. Grove TL, et al. Biochemistry. 2013 Apr 30;52(17):2874-87. doi: 10.1021/bi400136u. Epub 2013 Apr 16. Biochemistry. 2013. PMID: 23477283 Free PMC article. - Peptide Selenocysteine Substitutions Reveal Direct Substrate-Enzyme Interactions at Auxiliary Clusters in Radical _S_-Adenosyl-l-methionine Maturases.
Rush KW, Eastman KAS, Kincannon WM, Blackburn NJ, Bandarian V. Rush KW, et al. J Am Chem Soc. 2023 May 10;145(18):10167-10177. doi: 10.1021/jacs.3c00831. Epub 2023 Apr 27. J Am Chem Soc. 2023. PMID: 37104670 Free PMC article. - The thiostrepton A tryptophan methyltransferase TsrM catalyses a cob(II)alamin-dependent methyl transfer reaction.
Benjdia A, Pierre S, Gherasim C, Guillot A, Carmona M, Amara P, Banerjee R, Berteau O. Benjdia A, et al. Nat Commun. 2015 Oct 12;6:8377. doi: 10.1038/ncomms9377. Nat Commun. 2015. PMID: 26456915 Free PMC article.
References
- Hanson, S. R., Best, M. D., and Wong, C. H. (2004) Angew. Chem. Int. Ed. Engl. 43 5736–5763 - PubMed
- Szameit, C., Miech, C., Balleininger, M., Schmidt, B., von Figura, K., and Dierks, T. (1999) J. Biol. Chem. 274 15375–15381 - PubMed
- Fang, Q., Peng, J., and Dierks, T. (2004) J. Biol. Chem. 279 14570–14578 - PubMed
- Berteau, O., Guillot, A., Benjdia, A., and Rabot, S. (2006) J. Biol. Chem. 281 22464–22470 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources