CHIP deficiency decreases longevity, with accelerated aging phenotypes accompanied by altered protein quality control - PubMed (original) (raw)
CHIP deficiency decreases longevity, with accelerated aging phenotypes accompanied by altered protein quality control
Jin-Na Min et al. Mol Cell Biol. 2008 Jun.
Abstract
During the course of biological aging, there is a gradual accumulation of damaged proteins and a concomitant functional decline in the protein degradation system. Protein quality control is normally ensured by the coordinated actions of molecular chaperones and the protein degradation system that collectively help to maintain protein homeostasis. The carboxyl terminus of Hsp70-interacting protein (CHIP), a ubiquitin ligase/cochaperone, participates in protein quality control by targeting a broad range of chaperone substrates for proteasome degradation via the ubiquitin-proteasome system, demonstrating a broad involvement of CHIP in maintaining cytoplasmic protein quality control. In the present study, we have investigated the influence that protein quality control exerts on the aging process by using CHIP-/- mice. CHIP deficiency in mice leads to a markedly reduced life span, along with accelerated age-related pathophysiological phenotypes. These features were accompanied by indications of accelerated cellular senescence and increased indices of oxidative stress. In addition, CHIP-/- mice exhibit a deregulation of protein quality control, as indicated by elevated levels of toxic oligomer proteins and a decline in proteasome activity. Taken together, these data reveal that impaired protein quality control contributes to cellular senescence and implicates CHIP-dependent quality control mechanisms in the regulation of mammalian longevity in vivo.
Figures
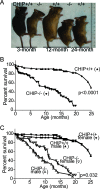
FIG. 1.
CHIP−/− mice have shortened life spans compared to those of CHIP+/+ mice. (A) Representative photographs of CHIP+/+ and CHIP−/− mice in indicated age groups, showing that CHIP−/− mice demonstrated small body sizes and enhanced kyphosis relative to age-matched CHIP+/+ mice. (B) Kaplan-Meier survival analyses in a cohort (male and female) of CHIP+/+ (n = 82) and CHIP−/− (n = 128) mice. CHIP−/− mice have significantly shortened life spans compared with those of CHIP+/+ mice (log rank test, P < 0.0001). (C) Kaplan-Meier survival analyses in CHIP+/+ male (n = 82), CHIP+/+ female (n = 84), CHIP−/− male (n = 58), and CHIP−/− female (n = 45) mouse groups. CHIP−/− male mice showed reduced longevity compared with that of CHIP−/− female mice (log rank test, P = 0.0324).
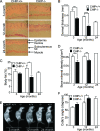
FIG. 2.
CHIP−/− mice exhibit a premature aging phenotype. (A) Representative histological sections of skin from CHIP−/− and CHIP+/+ mice at indicated age time points (original magnification, ×200). The black bar indicates the dermis layer within the skin section. (B) Quantification of dermal thickness from dorsal skin in indicated genotypes and age groups (three or four mice per group). (C and D) Quantification of body fat stores (C) and bone mineral density (D) measured by DEXA scan in indicated genotypes and age groups (four to eight mice per group). (E) Representative whole-body radiographs show the degree of kyphosis in CHIP−/− and CHIP+/+ mice in indicated age groups. (F) Quantification of Cobb's angle, which represents kyphosis, in indicated genotypes and age groups (three to five per group). Data shown are means ± standard errors of the means. Error bars indicate standard deviations. Asterisks indicate statistically significant differences (*, P < 0.05; **, P < 0.01; ***, P < 0.001 [Student's t test]).
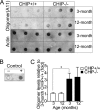
FIG. 3.
Deficiency of CHIP leads to increased SA-β-Gal activity and replicative senescence. (A) Photographs of SA-β-Gal staining in kidneys from CHIP−/− and CHIP+/+ mice in indicated age groups. SA-β-Gal-positive staining (blue) in the renal cortex was apparent in kidneys from CHIP−/− mice at 3 and 12 months of age as well as in kidneys from 24-month-old CHIP+/+ mice. (B) Representative images of SA-β-Gal staining in CHIP−/− and CHIP+/+ MEFs at P3 and P8 (original magnification, ×200). (C) Quantitative analysis of the percentage of SA-β-Gal-positive cells from CHIP−/− and CHIP+/+ MEFs at indicated passages. Data shown are means ± standard errors of the means. Error bars indicate standard deviations. P values were determined by Student's t test. (D) 3T9 proliferation analysis in CHIP−/− and CHIP+/+ primary MEFs from different passages. Data shown are mean ± standard deviations. P values were determined by Student's t test.
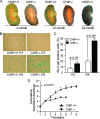
FIG. 4.
Accumulation of misfolded proteins in CHIP−/− mice. (A) Expression of misfolded oligomer proteins in brain tissues obtained from indicated genotypes and age groups, as detected by a dot blot analysis using antioligomer (A11) antibody. Actin was used as a loading control. (B) Dot blot analysis showing positive and negative controls for the antioligomer antibody (see Materials and Methods). (C) Quantification of oligomer expression in brain tissue from the dot blot shown in panel A. Expression of oligomer was measured relative to the oligomer level in brain tissues of 3-month-old CHIP+/+ mice. Data shown are means ± standard errors of the means. Error bars indicate standard deviations. *, P < 0.05 (Student's t test).
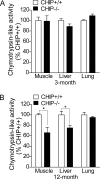
FIG. 5.
26S proteasome activity in CHIP−/− mice. Comparison of chymotrypsin-like activity in indicated tissues from CHIP−/− and CHIP+/+ mice at 3-month (A) and 12-month (B) age groups (three mice per group). Chymotrypsin-like activity was measured as described in Materials and Methods by using a fluorogenic substrate (Suc-LLVY-AMC) specific for chymotrypsin-like activity. The percentile of 26S proteasome activity was measured relative to the activity in CHIP+/+ mice from each age group. Data shown are means ± standard errors of the means. Error bars indicate standard deviations. *, P < 0.05 (Student's t test).
Similar articles
- Regulation of the cytoplasmic quality control protein degradation pathway by BAG2.
Dai Q, Qian SB, Li HH, McDonough H, Borchers C, Huang D, Takayama S, Younger JM, Ren HY, Cyr DM, Patterson C. Dai Q, et al. J Biol Chem. 2005 Nov 18;280(46):38673-81. doi: 10.1074/jbc.M507986200. Epub 2005 Sep 16. J Biol Chem. 2005. PMID: 16169850 - CHIP E3 ligase regulates mammalian senescence by modulating the levels of oxidized proteins.
Sisoula C, Gonos ES. Sisoula C, et al. Mech Ageing Dev. 2011 May;132(5):269-72. doi: 10.1016/j.mad.2011.04.003. Epub 2011 Apr 12. Mech Ageing Dev. 2011. PMID: 21510971 - Cooperation of a ubiquitin domain protein and an E3 ubiquitin ligase during chaperone/proteasome coupling.
Demand J, Alberti S, Patterson C, Höhfeld J. Demand J, et al. Curr Biol. 2001 Oct 16;11(20):1569-77. doi: 10.1016/s0960-9822(01)00487-0. Curr Biol. 2001. PMID: 11676916 - CHIP: a co-chaperone for degradation by the proteasome.
Edkins AL. Edkins AL. Subcell Biochem. 2015;78:219-42. doi: 10.1007/978-3-319-11731-7_11. Subcell Biochem. 2015. PMID: 25487024 Review. - CHIP: a quality-control E3 ligase collaborating with molecular chaperones.
Murata S, Chiba T, Tanaka K. Murata S, et al. Int J Biochem Cell Biol. 2003 May;35(5):572-8. doi: 10.1016/s1357-2725(02)00394-1. Int J Biochem Cell Biol. 2003. PMID: 12672450 Review.
Cited by
- CHIP protects against cardiac pressure overload through regulation of AMPK.
Schisler JC, Rubel CE, Zhang C, Lockyer P, Cyr DM, Patterson C. Schisler JC, et al. J Clin Invest. 2013 Aug;123(8):3588-99. doi: 10.1172/JCI69080. Epub 2013 Jul 25. J Clin Invest. 2013. PMID: 23863712 Free PMC article. - Uptake of Flaxseed Dietary Linusorbs Modulates Regulatory Genes Including Induction of Heat Shock Proteins and Apoptosis.
Shim YY, Tse TJ, Saini AK, Kim YJ, Reaney MJT. Shim YY, et al. Foods. 2022 Nov 22;11(23):3761. doi: 10.3390/foods11233761. Foods. 2022. PMID: 36496568 Free PMC article. - HSF1Base: A Comprehensive Database of HSF1 (Heat Shock Factor 1) Target Genes.
Kovács D, Sigmond T, Hotzi B, Bohár B, Fazekas D, Deák V, Vellai T, Barna J. Kovács D, et al. Int J Mol Sci. 2019 Nov 19;20(22):5815. doi: 10.3390/ijms20225815. Int J Mol Sci. 2019. PMID: 31752429 Free PMC article. - Maintenance of muscle mass and load-induced growth in Muscle RING Finger 1 null mice with age.
Hwee DT, Baehr LM, Philp A, Baar K, Bodine SC. Hwee DT, et al. Aging Cell. 2014 Feb;13(1):92-101. doi: 10.1111/acel.12150. Epub 2013 Sep 12. Aging Cell. 2014. PMID: 23941502 Free PMC article. - Cystic Fibrosis Transmembrane Conductance Regulator (CFTR) Ubiquitylation as a Novel Pharmaceutical Target for Cystic Fibrosis.
Fukuda R, Okiyoneda T. Fukuda R, et al. Pharmaceuticals (Basel). 2020 Apr 22;13(4):75. doi: 10.3390/ph13040075. Pharmaceuticals (Basel). 2020. PMID: 32331485 Free PMC article. Review.
References
- Ballinger, C. A., P. Connell, Y. Wu, Z. Hu, L. J. Thompson, L.-Y. Yin, and C. Patterson. 1999. Identification of CHIP, a novel tetratricopeptide repeat-containing protein that interacts with heat shock proteins and negatively regulates chaperone functions. Mol. Cell. Biol. 194535-4545. - PMC - PubMed
- Barral, J. M., S. A. Broadley, G. Schaffar, and F. U. Hartl. 2004. Roles of molecular chaperones in protein misfolding diseases. Semin. Cell Dev. Biol. 1517-29. - PubMed
- Bence, N. F., R. M. Sampat, and R. R. Kopito. 2001. Impairment of the ubiquitin-proteasome system by protein aggregation. Science 2921552-1555. - PubMed
- Bokov, A., A. Chaudhuri, and A. Richardson. 2004. The role of oxidative damage and stress in aging. Mech. Ageing Dev. 125811-826. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- HL65619/HL/NHLBI NIH HHS/United States
- R37 HL065619/HL/NHLBI NIH HHS/United States
- GM61728/GM/NIGMS NIH HHS/United States
- P01 AG024282/AG/NIA NIH HHS/United States
- AG024282/AG/NIA NIH HHS/United States
- R01 HL065619/HL/NHLBI NIH HHS/United States
- R01 GM061728/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases