Using genetic markers to orient the edges in quantitative trait networks: the NEO software - PubMed (original) (raw)
Using genetic markers to orient the edges in quantitative trait networks: the NEO software
Jason E Aten et al. BMC Syst Biol. 2008.
Abstract
Background: Systems genetic studies have been used to identify genetic loci that affect transcript abundances and clinical traits such as body weight. The pairwise correlations between gene expression traits and/or clinical traits can be used to define undirected trait networks. Several authors have argued that genetic markers (e.g expression quantitative trait loci, eQTLs) can serve as causal anchors for orienting the edges of a trait network. The availability of hundreds of thousands of genetic markers poses new challenges: how to relate (anchor) traits to multiple genetic markers, how to score the genetic evidence in favor of an edge orientation, and how to weigh the information from multiple markers.
Results: We develop and implement Network Edge Orienting (NEO) methods and software that address the challenges of inferring unconfounded and directed gene networks from microarray-derived gene expression data by integrating mRNA levels with genetic marker data and Structural Equation Model (SEM) comparisons. The NEO software implements several manual and automatic methods for incorporating genetic information to anchor traits. The networks are oriented by considering each edge separately, thus reducing error propagation. To summarize the genetic evidence in favor of a given edge orientation, we propose Local SEM-based Edge Orienting (LEO) scores that compare the fit of several competing causal graphs. SEM fitting indices allow the user to assess local and overall model fit. The NEO software allows the user to carry out a robustness analysis with regard to genetic marker selection. We demonstrate the utility of NEO by recovering known causal relationships in the sterol homeostasis pathway using liver gene expression data from an F2 mouse cross. Further, we use NEO to study the relationship between a disease gene and a biologically important gene co-expression module in liver tissue.
Conclusion: The NEO software can be used to orient the edges of gene co-expression networks or quantitative trait networks if the edges can be anchored to genetic marker data. R software tutorials, data, and supplementary material can be downloaded from: http://www.genetics.ucla.edu/labs/horvath/aten/NEO.
Figures
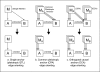
Figure 1
Approaches for genetic marker-based causal inference. Here we contrast different approaches for causality testing based on genetic markers. (a) single marker edge orienting involving a candidate pleiotropic anchor (CPA) M. The upper half of (a) shows the starting point of network edge orienting based on a single genetic marker M which is associated with traits A and B. The undirected edge between A and B indicates a significant correlation cor(A, B) between the two traits. The causal model in the lower half of (a) implies the following relationship between the correlation coefficients cor(M, B) = cor(M, A) × cor(A, B). Further it implies that the absolute value of the correlations |cor(M, A)| and |cor(M, B)| are high whereas the partial correlation |cor(M, B|A)| (Eq. 1) is low. Figure (b) generalizes the single marker situation to the case of multiple genetic markers MA={MA(1),MA(2),...}. In this case, it is straightforward to generalize single edge orienting scores to multi-marker scores. Figure (c) describes a situation when a set of genetic markers MB={MB(1),MB(2),...} is also available for trait B. We refer to the M B markers as orthogonal causal anchors (OCA) since cor(A,MB(j)) is expected to be 0 under the causal model M A → A → B → M B, the correlation. Using simulation studies, we find that edge scores based on OCAs can be more powerful than those based on CPAs (see Additional File 1).
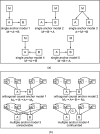
Figure 2
Illustrating the single genetic marker versus multi-marker local SEMs used in the definition of the LEO.NB score. The single genetic marker is denoted by M in (a) and the multiple genetic markers are denoted by MA(i) and MB(j) in (b) and (c). By definition, LEO.NB(A_˃_B) = log10{P (model 1)}/{max_i_>1{P (model i)}} for a candidate A → B edge orientation, where the models in the definition are pictured in (a) for single marker LEO.NB scores, and in (b) for multiple marker LEO.NB scores. In (b) we show the orthomarker models used for the LEO.NB.OCA marker aggregation method. The hidden confounder C in model 4 is the causal parent of both A and B, i.e. A ← C → B. The simulation studies in Additional File 1 show that the LEO.NB.OCA score can be significantly more powerful than the LEO.NB.CPA score.
Figure 3
Overview of the network edge orienting method. The steps of the network overview analysis are described in the text.
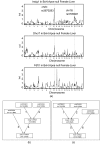
Figure 4
Manual SNP selection to study Insig1 → Dhrc7 and Insig1 → Fdft1 in mouse liver. Using female liver gene expression data and SNP markers from the BxH mouse intercross, NEO retrieves known causal relationships in the cholesterol biosynthesis pathway: Insig1 → Dhrc7 and Insig1 → Fdft1. The single marker LOD score curves in (a) motivate our choice of manually selected SNPs (one SNP on chromosome 16 and another on chromosome 8). These SNP markers can also be used to screen for genes that are reactive to Insig1, see Table 2. Figures (b) and (c) show the causal models used to compute the model p-values in favor of edge orientations _Insig_1 → _Dhcr_7 and _Insig_1 → _Fdft_1, respectively. More details on the individual edges are presented in Table 1.
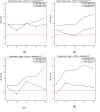
Figure 5
Automatic SNP selection to score Insig1 → Dhrc7 and Insig1 → Fdft1 in female and male mouse livers. These robustness plots show how the LEO.NB scores (y-axis) depend on sets of automatically selected SNP markers (x-axis). Here we use the default SNP selection method: combined greedy and forward stepwise method. Step K corresponds to choosing the top K greedy and top K forward selected SNPs for each trait. Since the greedy and the forward SNP selection may select the same SNPs, step K typically involves fewer than 2_K_ SNPs per trait. Figures (a, b, top row) and (c, d) correspond to female and male BxH mice, respectively. Figures (a) and (c) report the results for edge _Insig_1 → _Dhrc_7 in female and male mouse livers, respectively. Figures (b) and (d) report the analogous results for _Insig_1 → _Fdft_1. NEO robustly retrieves the known causal relationship between these genes.
Figure 6
Fsp27 is a causal driver of a biologically important co-expression module. Prior work using mouse liver expression data found the 'blue' co-expression module to be biologically important [7]. Here we used automatic SNP selection to determine whether Fsp27 is causal of the blue module gene expression profiles. The expression profiles of the blue module were summarized by their first principal component (referred to as module eigengene). The blue module eigengene MEblue can be considered as the most representative gene expression profile of the blue module. The figure shows the results of a robustness analysis regarding LEO.NB(_Fsp_27 → MEblue) (y-axis) with respect to different choices of genetic markers sets (x-axis). Both LEO.NB.CPA and LEO.NB.OCA scores show that the relationship is causal, i.e. the _Fsp_27 is upstream of the blue module expressions.
Figure 7
Multi-edge simulation study involving 5 gene expression traits (_E_1-_E_5) and one clinical trait Trait. The heatmap plot in (a) depicts the true causal model. Note that a red square in the i-th row and j-th column indicates that trait i causally affects trait j, e.g. _E_1 → _E_2. The rows and columns of the heatmap are ordered according to a hierarchical clustering tree, which was constructed using average linkage hierarchical clustering based on the pairwise correlations of the traits. Figure (b) depicts the corresponding heatmap of the observed network that was reconstructed using the LEO.NB.OCA score. Figure (c) shows an alternative output graph of NEO. Blue edges indicate significant correlations and a LEO.NB.OCA score is added to each edges whose LEO.NB.OCA score passes a user-supplied threshold. We find that all true causal edges are correctly retrieved at the recommended LEO.NB.OCA threshold of 0.3. Figure (d) shows the results of a robustness analysis for the LEO.NB.OCA and LEO.NB.CPA scores for the edge orientation _E_4 → Trait. The LEO.NB.OCA scores exceed the recommended threshold of 0.3 (red horizontal line), i.e. they retrieve the orientation correctly. Similarly, the LEO.NB.CPA scores exceed the threshold of 0.8.
Similar articles
- FunMap: functional mapping of complex traits.
Ma CX, Wu R, Casella G. Ma CX, et al. Bioinformatics. 2004 Jul 22;20(11):1808-11. doi: 10.1093/bioinformatics/bth156. Epub 2004 Feb 26. Bioinformatics. 2004. PMID: 14988108 - R/qtl: QTL mapping in experimental crosses.
Broman KW, Wu H, Sen S, Churchill GA. Broman KW, et al. Bioinformatics. 2003 May 1;19(7):889-90. doi: 10.1093/bioinformatics/btg112. Bioinformatics. 2003. PMID: 12724300 - Eigengene networks for studying the relationships between co-expression modules.
Langfelder P, Horvath S. Langfelder P, et al. BMC Syst Biol. 2007 Nov 21;1:54. doi: 10.1186/1752-0509-1-54. BMC Syst Biol. 2007. PMID: 18031580 Free PMC article. - eQTL analysis in mice and rats.
Tesson BM, Jansen RC. Tesson BM, et al. Methods Mol Biol. 2009;573:285-309. doi: 10.1007/978-1-60761-247-6_16. Methods Mol Biol. 2009. PMID: 19763934 Review. - Moving toward a system genetics view of disease.
Sieberts SK, Schadt EE. Sieberts SK, et al. Mamm Genome. 2007 Jul;18(6-7):389-401. doi: 10.1007/s00335-007-9040-6. Epub 2007 Jul 26. Mamm Genome. 2007. PMID: 17653589 Free PMC article. Review.
Cited by
- Cross-species transcriptional network analysis reveals conservation and variation in response to metal stress in cyanobacteria.
Wang J, Wu G, Chen L, Zhang W. Wang J, et al. BMC Genomics. 2013 Feb 19;14:112. doi: 10.1186/1471-2164-14-112. BMC Genomics. 2013. PMID: 23421563 Free PMC article. - The genetics of quantitative traits: challenges and prospects.
Mackay TF, Stone EA, Ayroles JF. Mackay TF, et al. Nat Rev Genet. 2009 Aug;10(8):565-77. doi: 10.1038/nrg2612. Nat Rev Genet. 2009. PMID: 19584810 Review. - Genetical Genomics of Behavior: A Novel Chicken Genomic Model for Anxiety Behavior.
Johnsson M, Williams MJ, Jensen P, Wright D. Johnsson M, et al. Genetics. 2016 Jan;202(1):327-40. doi: 10.1534/genetics.115.179010. Genetics. 2016. PMID: 26733665 Free PMC article. - Gene regulatory network reconstruction using Bayesian networks, the Dantzig Selector, the Lasso and their meta-analysis.
Vignes M, Vandel J, Allouche D, Ramadan-Alban N, Cierco-Ayrolles C, Schiex T, Mangin B, de Givry S. Vignes M, et al. PLoS One. 2011;6(12):e29165. doi: 10.1371/journal.pone.0029165. Epub 2011 Dec 27. PLoS One. 2011. PMID: 22216195 Free PMC article. - Genetical Genomics of Tonic Immobility in the Chicken.
Fogelholm J, Inkabi S, Höglund A, Abbey-Lee R, Johnsson M, Jensen P, Henriksen R, Wright D. Fogelholm J, et al. Genes (Basel). 2019 May 7;10(5):341. doi: 10.3390/genes10050341. Genes (Basel). 2019. PMID: 31067744 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
- DGE9987641/PHS HHS/United States
- HL28481/HL/NHLBI NIH HHS/United States
- T32 HG002536/HG/NHGRI NIH HHS/United States
- P01 HL028481/HL/NHLBI NIH HHS/United States
- U19 AI063603/AI/NIAID NIH HHS/United States
- HG02536-04/HG/NHGRI NIH HHS/United States
- HL30568/HL/NHLBI NIH HHS/United States
- P01 HL030568/HL/NHLBI NIH HHS/United States
- 1U19AI063603-01/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous