Small RNA guides for de novo DNA methylation in mammalian germ cells - PubMed (original) (raw)
Comment
Small RNA guides for de novo DNA methylation in mammalian germ cells
Alexei A Aravin et al. Genes Dev. 2008.
Abstract
Germline genomic methylation is essential for gamete identity and integrity in mammals. The study by Kuramochi-Miyagawa and colleagues (908-917) in the previous issue of Genes & Development links the process of DNA methylation-dependent repression of retrotranspons with the presence of piwi-interacting RNAs (piRNAs) in fetal male germ cells undergoing de novo methylation.
Figures
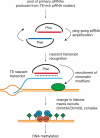
Figure 1.
Proposed mechanism for piRNA-induced DNA methylation in male germ cells. Primary piRNAs produced from TE-rich piRNA clusters join Piwi (Mili and/or Miwi2) proteins. Sense (blue) and antisense (red) piRNAs are amplified in the ping-pong cycle. Piwi–piRNAs complexes recognize nascent TE transcripts in the nucleus and recruit chromatin modifiers to TE genomic loci. Subsequent changes in histone marks (H3K4 methylation loss, H3K9 methylation gain, green diamonds) induce the de novo activity of the Dnmt3A–Dnmt3L tetrameric complex to establish methylation patterns.
Comment on
- DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes.
Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Totoki Y, Toyoda A, Ikawa M, Asada N, Kojima K, Yamaguchi Y, Ijiri TW, Hata K, Li E, Matsuda Y, Kimura T, Okabe M, Sakaki Y, Sasaki H, Nakano T. Kuramochi-Miyagawa S, et al. Genes Dev. 2008 Apr 1;22(7):908-17. doi: 10.1101/gad.1640708. Genes Dev. 2008. PMID: 18381894 Free PMC article.
Similar articles
- Induction of DNA methylation by artificial piRNA production in male germ cells.
Itou D, Shiromoto Y, Yukiho SY, Ishii C, Nishimura T, Ogonuki N, Ogura A, Hasuwa H, Fujihara Y, Kuramochi-Miyagawa S, Nakano T. Itou D, et al. Curr Biol. 2015 Mar 30;25(7):901-6. doi: 10.1016/j.cub.2015.01.060. Epub 2015 Mar 12. Curr Biol. 2015. PMID: 25772451 - PIWI-interacting RNAs (piRNAs) - a mouse testis perspective.
Bortvin A. Bortvin A. Biochemistry (Mosc). 2013 Jun;78(6):592-602. doi: 10.1134/S0006297913060059. Biochemistry (Mosc). 2013. PMID: 23980886 Review. - MIWI2 as an Effector of DNA Methylation and Gene Silencing in Embryonic Male Germ Cells.
Kojima-Kita K, Kuramochi-Miyagawa S, Nagamori I, Ogonuki N, Ogura A, Hasuwa H, Akazawa T, Inoue N, Nakano T. Kojima-Kita K, et al. Cell Rep. 2016 Sep 13;16(11):2819-2828. doi: 10.1016/j.celrep.2016.08.027. Cell Rep. 2016. PMID: 27626653 - Switching of dominant retrotransposon silencing strategies from posttranscriptional to transcriptional mechanisms during male germ-cell development in mice.
Inoue K, Ichiyanagi K, Fukuda K, Glinka M, Sasaki H. Inoue K, et al. PLoS Genet. 2017 Jul 27;13(7):e1006926. doi: 10.1371/journal.pgen.1006926. eCollection 2017 Jul. PLoS Genet. 2017. PMID: 28749988 Free PMC article. - Small RNA silencing pathways in germ and stem cells.
Aravin AA, Hannon GJ. Aravin AA, et al. Cold Spring Harb Symp Quant Biol. 2008;73:283-90. doi: 10.1101/sqb.2008.73.058. Epub 2009 Mar 6. Cold Spring Harb Symp Quant Biol. 2008. PMID: 19270082 Review.
Cited by
- Human Endogenous Retroviruses in Diseases.
Fan TJ, Cui J. Fan TJ, et al. Subcell Biochem. 2023;106:403-439. doi: 10.1007/978-3-031-40086-5_15. Subcell Biochem. 2023. PMID: 38159236 - Epigenetic Reprogramming and Somatic Cell Nuclear Transfer.
Vargas LN, Silveira MM, Franco MM. Vargas LN, et al. Methods Mol Biol. 2023;2647:37-58. doi: 10.1007/978-1-0716-3064-8_2. Methods Mol Biol. 2023. PMID: 37041328 Review. - Finer resolution analysis of transcriptional programming during the active migration of chicken primordial germ cells.
Rengaraj D, Cha DG, Park KJ, Lee KY, Woo SJ, Han JY. Rengaraj D, et al. Comput Struct Biotechnol J. 2022 Oct 26;20:5911-5924. doi: 10.1016/j.csbj.2022.10.034. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 36382185 Free PMC article. - Toxicoepigenetics and Environmental Health: Challenges and Opportunities.
Svoboda LK, Perera BPU, Morgan RK, Polemi KM, Pan J, Dolinoy DC. Svoboda LK, et al. Chem Res Toxicol. 2022 Aug 15;35(8):1293-1311. doi: 10.1021/acs.chemrestox.1c00445. Epub 2022 Jul 25. Chem Res Toxicol. 2022. PMID: 35876266 Free PMC article. Review. - Epigenetic Regulation in Hydra: Conserved and Divergent Roles.
Pillai A, Gungi A, Reddy PC, Galande S. Pillai A, et al. Front Cell Dev Biol. 2021 May 10;9:663208. doi: 10.3389/fcell.2021.663208. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34041242 Free PMC article. Review.
References
- Aravin A.A., Lagos-Quintana M., Yalcin A., Zavolan M., Marks D., Snyder B., Gaasterland T., Meyer J., Tuschl T. The small RNA profile during Drosophilia melanogaster development. Dev. Cell. 2003;5:337–350. - PubMed
- Aravin A., Gaidatzis D., Pfeffer S., Lagos-Quintana M., Landgraf P., Iovino N., Morris P., Brownstein M.J., Kuramochi-Miyagawa S., Nakano T., et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. - PubMed
- Aravin A.A., Sachidanandam R., Girard A., Fejes-Toth K., Hannon G.J. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007a;316:744–747. - PubMed
- Aravin A.A., Hannon G.J., Brennecke J. The Piwi–piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007b;318:761–764. - PubMed
- Barlow D.P. Methylation and imprinting: From host defense to gene regulation? Science. 1993;260:309–310. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources