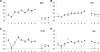Host PrP glycosylation: a major factor determining the outcome of prion infection - PubMed (original) (raw)
Host PrP glycosylation: a major factor determining the outcome of prion infection
Nadia L Tuzi et al. PLoS Biol. 2008.
Abstract
The expression of the prion protein (PrP) is essential for transmissible spongiform encephalopathy (TSE) or prion diseases to occur, but the underlying mechanism of infection remains unresolved. To address the hypothesis that glycosylation of host PrP is a major factor influencing TSE infection, we have inoculated gene-targeted transgenic mice that have restricted N-linked glycosylation of PrP with three TSE strains. We have uniquely demonstrated that mice expressing only unglycosylated PrP can sustain a TSE infection, despite altered cellular location of the host PrP. Moreover we have shown that brain material from mice infected with TSE that have only unglycosylated PrP(Sc) is capable of transmitting infection to wild-type mice, demonstrating that glycosylation of PrP is not essential for establishing infection within a host or for transmitting TSE infectivity to a new host. We have further dissected the requirement of each glycosylation site and have shown that different TSE strains have dramatically different requirements for each of the glycosylation sites of host PrP, and moreover, we have shown that the host PrP has a major role in determining the glycosylation state of de novo generated PrP(Sc).
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures
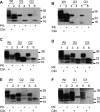
Figure 1. Western Blot Analysis of Brains from TSE Inoculated Glycosylation Mutant Mice
Brain homogenates were treated with or without PK as indicated prior to SDS PAGE analysis. (A) G3 and wild-type animals inoculated with 79A. Lanes 1–2: 129/Ola wild-type mice clinically (clin) positive/pathologically (path) positive; lanes 3–4: G3 mice clin positive/path positive; lanes 5–6: G3 mice clin negative/path positive. (B) G3 and wild-type animals inoculated with ME7. Lanes 1–2: 129/Ola mice clin positive/path positive; lanes 3–4: G3 mice clin negative/path negative with no plaques; lanes 5–6: G3 mice clin negative/path negative with plaques. (C) G2 and wild-type animals inoculated with ME7. Lanes 1–2: 129/Ola; lanes 3–6: G2 mice, brains positive for plaques. All animals clin positive/path positive. (D) G2 and wild-type animals inoculated with 79A. Lanes 1–2: 129/Ola mice; lanes 3–6: G2 mice. All animals clin positive/path positive. (E) G2 and wild-type animals inoculated with 301C. Lanes 1–2: 129/Ola mice; lanes 3–6: G2 mice brains positive for plaques. All animals clin positive/path positive. (F) G1 and wild-type animals inoculated with 79A. Lanes 1–2: 129/Ola mice; lanes 3–6: G1 mice. All animals clin positive/path positive. Lanes 1–4: 3-min exposure; lanes 5–6: 10-min exposure. Molecular weight markers indicated in KDa. Wild-type (Wt) mice are 129/Ola mice. Monoclonal antibody 7A12 (kindly provided by M.-S. Sy) was used for immunodetection of PrP.
Figure 2. Immunohistochemical Analysis of Brain from G3 Mice Inoculated with TSE Strains 79A or ME7
Sections from the cerebrum and cerebellum were immunostained for PrP with monoclonal antibody 6H4 and analysed by light microscopy using a Nikon Eclipse E 800 microscope. Brain sections obtained from (A) clinically negative, or (B) clinically positive G3 mice after inoculation with 79A showing widespread accumulation of PrP in the hippocampus and thalamus are shown. (C and D) Brain sections from clinically negative G3 mice inoculated with ME7 showing large PrP positive plaques in subcallosal areas. (E and F) Brain section of clinically positive 129/Ola mice infected with 79A (E) or ME7 (F), showing widespread PrP deposition in the hippocampus and thalamus. (G) Thioflavin-S fluorescent PrP-amyloid plaques in the subcallosal region of a clinically negative G3 mouse inoculated with 79A. (H) Thioflavin-S treatment of sections from a clinically positive 129/Ola mouse infected with 79A with no presence of plaques. (I) Thioflavin-S fluorescent PrP-amyloid plaques in a clinically negative G3 mouse infected with ME7. (J) Thioflavin-S staining of a clinically positive 129/Ola mouse infected with ME7 with no presence of plaques. Magnifications: (A), (B), (E), and (F): 4×, (C), (G), (H), and (I): 10×; and (D): 40× .
Figure 3. Lesion Profile Analysis of TSE-Inoculated PrP Glycosylation Mutant Mice
G2 and 129/Ola mice were inoculated i.c. with TSE strains ME7 (A), 79A (B), or 301C (C) and G1 mice were inoculated i.c. with 79A (D). Squares: wild type; open triangles: G2; and solid triangles: G1. All mice (12 in each group) used in these analyses were clinically and pathologically positive. Nine gray matter areas: 1, dorsal medulla; 2, cerebellar cortex; 3, superior colliculus; 4, hypothalamus; 5, medial thalamus; 6, hippocampus; 7, septum; 8, cerebral cortex; 9, forebrain cortex and three white matter areas: 1*, cerebellar white matter; 2*, mesencephalic tegmentum; 3*, pyramidal tract (_x_-axis) were scored on scales of 0–5 for gray and 0–3 for white matter areas (_y_-axis). The mean scores for each area are shown (error bars ± SEM). No differences between wild-type and G2 mice were observed after inoculation with ME7 (A) or 79A (B); however some differences in the superior colliculus and white matter areas were observed between the two groups when inoculated with 301C (C). Differences were observed between wild-type and G1 mice inoculated with 79A (D) especially in area: 2, 4, 8, and in the white matter.
Figure 4. Immunohistochemical Analysis of Brain from G2 Mice Inoculated with TSE Strains ME7, 79A, and 301C
Brain sections were immunostained for PrP using monoclonal antibody 6H4 and analysed by light microscopy using a Nikon Eclipse E 800 microscope. Brains from clinically positive G2 mice inoculated with (A) ME7 or (B) 79A showing wide spread PrP deposition. Brains from 129/Ola wild-type mice inoculated with (C) ME7 or (D) 79A also showing wide spread PrP deposition. The TSE strain 301C was used to inoculate (E and F) G2 mice, which showed wide spread plaque-like and fine punctuate PrP deposits in the cerebrum and cerebellum or (G and H) 129Ola mice, which also showed plaque-like PrP deposition. All 301C inoculated brains shown were from clinically positive mice. Magnifications: (A–E) and (G), 4×; (F) and (H), 10×.
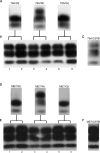
Figure 5. Western Blot Analysis of Brains from Second Passage of 79A-Infected G2 and Wild-Type Mice
Brain homogenates from (A) clinically and pathologically positive 79A inoculated G2 or wild-type, 129/Ola (Wt), mice were used to i.c. inoculate 129/Ola mice. (B) The 79A inoculated G2 mouse brains lacked diglycosylated PrPSc, but all 129/Ola mice inoculated with 79A/G2 or 79A/Wt brain material had di-, mono-, and unglycosylated PrPSc. (C) Brain of a C57Bl mouse infected with 79A used as comparison for the glycotype. Brain homogenates from (D) clinically and pathologically positive ME7 inoculated-G2 or wild-type, 129/Ola (Wt), mice were used to intracerebrally inoculate 129/Ola mice. (E) The ME7 inoculated G2 mouse brains lacked diglycosylated PrPSc but all 129/Ola mice inoculated with ME7/G2 or ME7/Wt brain material had di-, mono-, and unglycosylated PrPSc. (F) Brain of a C57Bl mouse infected with ME7 used as comparison for the glycotype. Lanes 1–6: TSE-inoculated 129/Ola mouse brain. All samples were PK treated.
Figure 6. Immunohistochemical Analysis of Brain from G1 Mice Inoculated with TSE Strains 79A and ME7
Brain sections were immunostained for PrP using monoclonal antibody 6H4 and analysed by light microscopy using a Nikon Eclipse E 800 microscope. Widespread PrP immunopositivity is seen mostly in the thalamus (A and C) of clinically positive, symptomatic, G1 mice inoculated with 79A, absence of PrP-immunopositivity in the hippocampus (B) and thalamus (D) of clinically (Clin) negative, asymptomatic, G1 mice inoculated with ME7, (E) widespread PrP deposition in the thalamus and hippocampus of a wild-type mouse (129/Ola) after inoculation with 79A; (F) widespread PrP deposition in the thalamus of a wild-type mouse with neurological signs after inoculation with ME7. Magnifications: (A), (B), (E), and (F), 4×; (C) and (D), 10×.
Similar articles
- Glycosylation of PrPC determines timing of neuroinvasion and targeting in the brain following transmissible spongiform encephalopathy infection by a peripheral route.
Cancellotti E, Bradford BM, Tuzi NL, Hickey RD, Brown D, Brown KL, Barron RM, Kisielewski D, Piccardo P, Manson JC. Cancellotti E, et al. J Virol. 2010 Apr;84(7):3464-75. doi: 10.1128/JVI.02374-09. Epub 2010 Jan 27. J Virol. 2010. PMID: 20106922 Free PMC article. - Post-translational changes to PrP alter transmissible spongiform encephalopathy strain properties.
Cancellotti E, Mahal SP, Somerville R, Diack A, Brown D, Piccardo P, Weissmann C, Manson JC. Cancellotti E, et al. EMBO J. 2013 Mar 6;32(5):756-69. doi: 10.1038/emboj.2013.6. Epub 2013 Feb 8. EMBO J. 2013. PMID: 23395905 Free PMC article. - Host and transmissible spongiform encephalopathy agent strain control glycosylation of PrP.
Somerville RA. Somerville RA. J Gen Virol. 1999 Jul;80 ( Pt 7):1865-1872. doi: 10.1099/0022-1317-80-7-1865. J Gen Virol. 1999. PMID: 10423157 - The role of host PrP in Transmissible Spongiform Encephalopathies.
Cancellotti E, Barron RM, Bishop MT, Hart P, Wiseman F, Manson JC. Cancellotti E, et al. Biochim Biophys Acta. 2007 Jun;1772(6):673-80. doi: 10.1016/j.bbadis.2006.10.013. Epub 2006 Oct 26. Biochim Biophys Acta. 2007. PMID: 17150338 Review. - Evolving views in prion glycosylation: functional and pathological implications.
Ermonval M, Mouillet-Richard S, Codogno P, Kellermann O, Botti J. Ermonval M, et al. Biochimie. 2003 Jan-Feb;85(1-2):33-45. doi: 10.1016/s0300-9084(03)00040-3. Biochimie. 2003. PMID: 12765773 Review.
Cited by
- Further Characterization of Glycoform-Selective Prions of Variably Protease-Sensitive Prionopathy.
Zhang W, Xiao X, Ding M, Yuan J, Foutz A, Moudjou M, Kitamoto T, Langeveld JPM, Cui L, Zou WQ. Zhang W, et al. Pathogens. 2021 Apr 23;10(5):513. doi: 10.3390/pathogens10050513. Pathogens. 2021. PMID: 33922765 Free PMC article. - Cell biology of prion strains in vivo and in vitro.
Shoup D, Priola SA. Shoup D, et al. Cell Tissue Res. 2023 Apr;392(1):269-283. doi: 10.1007/s00441-021-03572-y. Epub 2022 Feb 2. Cell Tissue Res. 2023. PMID: 35107622 Free PMC article. Review. - Getting a grip on prions: oligomers, amyloids, and pathological membrane interactions.
Caughey B, Baron GS, Chesebro B, Jeffrey M. Caughey B, et al. Annu Rev Biochem. 2009;78:177-204. doi: 10.1146/annurev.biochem.78.082907.145410. Annu Rev Biochem. 2009. PMID: 19231987 Free PMC article. Review. - Underglycosylated prion protein modulates plaque formation in the brain.
Bartz JC. Bartz JC. J Clin Invest. 2020 Mar 2;130(3):1087-1089. doi: 10.1172/JCI134842. J Clin Invest. 2020. PMID: 31985491 Free PMC article. - Biological Functions of the Intrinsically Disordered N-Terminal Domain of the Prion Protein: A Possible Role of Liquid-Liquid Phase Separation.
Polido SA, Kamps J, Tatzelt J. Polido SA, et al. Biomolecules. 2021 Aug 12;11(8):1201. doi: 10.3390/biom11081201. Biomolecules. 2021. PMID: 34439867 Free PMC article. Review.
References
- Manson JC, Clarke AR, McBride PA, McConnell I, Hope J. PrP gene dosage determines the timing but not the final intensity or distribution of lesions in scrapie pathology. Neurodegeneration. 1994;3:331–340. - PubMed
- DeArmond SJ, Sanchez H, Yehiely F, Qiu Y, Ninchak-Casey A, et al. Selective neuronal targeting in prion disease. Neuron. 1997;19:1337–1348. - PubMed
- Somerville RA, Chong A, Mulqueen OU, Birkett CR, Wood SC, et al. Biochemical typing of scrapie strains. Nature. 1997;386:564. - PubMed
- Safar J, Wille H, Itri V, Groth D, Serban H, et al. Eight prion strains have PrP(Sc)molecules with different conformations. Nat Med. 1998;4:1157–1165. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- BBS/E/A/00001655/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/D/05241340/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials