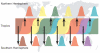The genomic and epidemiological dynamics of human influenza A virus - PubMed (original) (raw)
. 2008 May 29;453(7195):615-9.
doi: 10.1038/nature06945. Epub 2008 Apr 16.
Affiliations
- PMID: 18418375
- PMCID: PMC2441973
- DOI: 10.1038/nature06945
The genomic and epidemiological dynamics of human influenza A virus
Andrew Rambaut et al. Nature. 2008.
Abstract
The evolutionary interaction between influenza A virus and the human immune system, manifest as 'antigenic drift' of the viral haemagglutinin, is one of the best described patterns in molecular evolution. However, little is known about the genome-scale evolutionary dynamics of this pathogen. Similarly, how genomic processes relate to global influenza epidemiology, in which the A/H3N2 and A/H1N1 subtypes co-circulate, is poorly understood. Here through an analysis of 1,302 complete viral genomes sampled from temperate populations in both hemispheres, we show that the genomic evolution of influenza A virus is characterized by a complex interplay between frequent reassortment and periodic selective sweeps. The A/H3N2 and A/H1N1 subtypes exhibit different evolutionary dynamics, with diverse lineages circulating in A/H1N1, indicative of weaker antigenic drift. These results suggest a sink-source model of viral ecology in which new lineages are seeded from a persistent influenza reservoir, which we hypothesize to be located in the tropics, to sink populations in temperate regions.
Figures
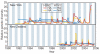
Figure 1. Population dynamics of genetic diversity in influenza A virus
Bayesian skyline plots of the HA and NA segments for the A/H3N2 and A/H1N1 subtypes in New York state (top) and New Zealand (bottom). The horizontal shaded blocks represent the winter seasons. The _y_-axes represent a measure of relative genetic diversity (see Methods for details). The shorter timescale of New Zealand skyline plot is due to the shorter sampling period.
Figure 2. A ‘source-sink’ model for the evolution of influenza A virus
Viral genetic and antigenic diversity (shown by different colours) is continuously generated in a reservoir, or ‘source’ population, perhaps represented by the tropics, before being exported to ‘sink’ populations in the Northern and Southern Hemispheres as shown by the arrows. The continuous transmission of influenza A virus in the source population, and hence its larger effective population size, allows natural selection for antigenic diversity to proceed more efficiently than in the sink populations that are afflicted by major seasonal bottlenecks.
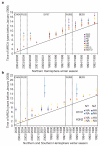
Figure 3. Population genetic history of human influenza A virus
a, b, Time to the most recent common ancestor (TMRCA) of each genomic segment for A/H3N2 isolates circulating each season in New York state (a) and HA and NA genomic segments for the A/H3N2 and A/H1N1 isolates circulating in New York state and New Zealand (b). The values shown represent the mean and 95% highest posterior density intervals for TMRCAs estimated across the trees sampled using bayesian MCMC analyses. Explicit posterior probabilities of whether any season has a TMRCA that is older than that of each preceding season are given in Supplementary Table 1. The diagonal line goes through 1st January of each season (1st July for New Zealand), approximating the seasonal mid-point. The timescale of major changes in antigenic (HA) type in the United States is also depicted. In the Northern Hemisphere, individual influenza seasons straddle two years, whereas in the Southern Hemisphere they are contained within one calendar year.
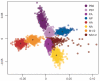
Figure 4. Differences in phylogenetic history among segments of H3N2 influenza A virus
A multi-dimensional scaling plot of distances between samples of 500 trees for each segment of A/H3N2 viruses sampled from New York state. The tree-to-tree distances are calculated as one minus the correlation coefficient of the vector of TMRCAs for each season. Each cloud of points of equal colour represents the statistical uncertainty in the phylogenetic history of an individual genomic segment (with the centroids of each segment distribution indicated). Considering each comparison in a pairwise manner between segments reveals a similar pattern (Supplementary Fig. 12).
Similar articles
- Genetic analysis of influenza A/H3N2 and A/H1N1 viruses circulating in Vietnam from 2001 to 2006.
Li D, Saito R, Le MT, Nguyen HL, Suzuki Y, Shobugawa Y, Dinh DT, Hoang PV, Tran HT, Nghiem HK, Hoang LT, Huynh LP, Nguyen HT, Nishikawa M, Suzuki H. Li D, et al. J Clin Microbiol. 2008 Feb;46(2):399-405. doi: 10.1128/JCM.01549-07. Epub 2007 Oct 17. J Clin Microbiol. 2008. PMID: 17942644 Free PMC article. - Genomewide analysis of reassortment and evolution of human influenza A(H3N2) viruses circulating between 1968 and 2011.
Westgeest KB, Russell CA, Lin X, Spronken MI, Bestebroer TM, Bahl J, van Beek R, Skepner E, Halpin RA, de Jong JC, Rimmelzwaan GF, Osterhaus AD, Smith DJ, Wentworth DE, Fouchier RA, de Graaf M. Westgeest KB, et al. J Virol. 2014 Mar;88(5):2844-57. doi: 10.1128/JVI.02163-13. Epub 2013 Dec 26. J Virol. 2014. PMID: 24371052 Free PMC article. - The evolution of human influenza A viruses from 1999 to 2006: a complete genome study.
Bragstad K, Nielsen LP, Fomsgaard A. Bragstad K, et al. Virol J. 2008 Mar 7;5:40. doi: 10.1186/1743-422X-5-40. Virol J. 2008. PMID: 18325125 Free PMC article. - [Swine influenza virus: evolution mechanism and epidemic characterization--a review].
Qi X, Lu C. Qi X, et al. Wei Sheng Wu Xue Bao. 2009 Sep;49(9):1138-45. Wei Sheng Wu Xue Bao. 2009. PMID: 20030049 Review. Chinese. - Mutation and Epistasis in Influenza Virus Evolution.
Lyons DM, Lauring AS. Lyons DM, et al. Viruses. 2018 Aug 3;10(8):407. doi: 10.3390/v10080407. Viruses. 2018. PMID: 30081492 Free PMC article. Review.
Cited by
- Genetic evolution of the neuraminidase of influenza A (H3N2) viruses from 1968 to 2009 and its correspondence to haemagglutinin evolution.
Westgeest KB, de Graaf M, Fourment M, Bestebroer TM, van Beek R, Spronken MIJ, de Jong JC, Rimmelzwaan GF, Russell CA, Osterhaus ADME, Smith GJD, Smith DJ, Fouchier RAM. Westgeest KB, et al. J Gen Virol. 2012 Sep;93(Pt 9):1996-2007. doi: 10.1099/vir.0.043059-0. Epub 2012 Jun 20. J Gen Virol. 2012. PMID: 22718569 Free PMC article. - Evolution of the H9N2 influenza genotype that facilitated the genesis of the novel H7N9 virus.
Pu J, Wang S, Yin Y, Zhang G, Carter RA, Wang J, Xu G, Sun H, Wang M, Wen C, Wei Y, Wang D, Zhu B, Lemmon G, Jiao Y, Duan S, Wang Q, Du Q, Sun M, Bao J, Sun Y, Zhao J, Zhang H, Wu G, Liu J, Webster RG. Pu J, et al. Proc Natl Acad Sci U S A. 2015 Jan 13;112(2):548-53. doi: 10.1073/pnas.1422456112. Epub 2014 Dec 29. Proc Natl Acad Sci U S A. 2015. PMID: 25548189 Free PMC article. - Reassortment with dominant chicken H9N2 influenza virus contributed to the fifth H7N9 virus human epidemic.
Pu J, Yin Y, Liu J, Wang X, Zhou Y, Wang Z, Sun Y, Sun H, Li F, Song J, Qu R, Gao W, Wang D, Wang Z, Yan S, Chen M, Zeng J, Jiang Z, Sun H, Zong Y, Wang C, Tong Q, Bi Y, Huang Y, Du X, Chang KC, Liu J. Pu J, et al. J Virol. 2021 May 10;95(11):e01578-20. doi: 10.1128/JVI.01578-20. Epub 2021 Mar 17. J Virol. 2021. PMID: 33731452 Free PMC article. - Molecular epidemiology of influenza A/H3N2 viruses circulating in Uganda.
Byarugaba DK, Ducatez MF, Erima B, Mworozi EA, Millard M, Kibuuka H, Lukwago L, Bwogi J, Kaira BB, Mimbe D, Schnabel DC, Krauss S, Darnell D, Webby RJ, Webster RG, Wabwire-Mangen F. Byarugaba DK, et al. PLoS One. 2011;6(11):e27803. doi: 10.1371/journal.pone.0027803. Epub 2011 Nov 21. PLoS One. 2011. PMID: 22132146 Free PMC article. - The age distribution of mortality due to influenza: pandemic and peri-pandemic.
Reichert T, Chowell G, McCullers JA. Reichert T, et al. BMC Med. 2012 Dec 12;10:162. doi: 10.1186/1741-7015-10-162. BMC Med. 2012. PMID: 23234604 Free PMC article.
References
- Drummond AJ, Rambaut A, Shapiro B, Pybus OG. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol. Biol. Evol. 2005;22:1185–1192. - PubMed
- Kingman J. The coalescent. Stochastic Process. Appl. 1982;13:235–248.
- Rambaut A. Estimating the rate of molecular evolution: Incorporating noncontemporaneous sequences into maximum likelihood phylogenies. Bioinformatics. 2000;16:395–399. - PubMed
- Shapiro B, Rambaut A, Drummond AJ. Choosing appropriate substitution models for the phylogenetic analysis of protein-coding sequences. Mol. Biol. Evol. 2006;23:7–9. - PubMed
- WHO Fact sheet Number 211. Influenza. 2003. http://www.who.int/mediacentre/factsheets/fs211/)
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical