O-GlcNAc regulates FoxO activation in response to glucose - PubMed (original) (raw)
O-GlcNAc regulates FoxO activation in response to glucose
Michael P Housley et al. J Biol Chem. 2008.
Abstract
FoxO proteins are key transcriptional regulators of nutrient homeostasis and stress response. The transcription factor FoxO1 activates expression of gluconeogenic, including phosphoenolpyruvate carboxykinase and glucose-6-phosphatase, and also activates the expression of the oxidative stress response enzymes catalase and manganese superoxide dismutase. Hormonal and stress-dependent regulation of FoxO1 via acetylation, ubiquitination, and phosphorylation, are well established, but FoxOs have not been studied in the context of the glucose-derived O-linked beta-N-acetylglucosamine (O-GlcNAc) modification. Here we show that O-GlcNAc on hepatic FoxO1 is increased in diabetes. Furthermore, O-GlcNAc regulates FoxO1 activation in response to glucose, resulting in the paradoxically increased expression of gluconeogenic genes while concomitantly inducing expression of genes encoding enzymes that detoxify reactive oxygen species. GlcNAcylation of FoxO provides a new mechanism for direct nutrient control of transcription to regulate metabolism and stress response through control of FoxO1 activity.
Figures
FIGURE 1.
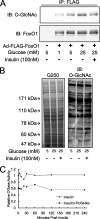
FoxO1 is _O-_GlcNAcylated. A, Fao cells were infected with Ad-FLAG-FoxO1. FLAG-FoxO1 was immunoprecipitated (IP) and blotted (IB) with the anti-O-GlcNAc antibody CTD110.6 or the terminal GlcNAc-binding lectin succinylated wheat germ agglutinin. Specificity was confirmed by GlcNAc competition and increased antibody or lectin reactivity following treatment with the _O-_GlcNAcase inhibitor PUGNAc.B, autoradiograph showing endogenous FoxO1 immunoprecipitated from rat liver and labeled with [3H]UDP-galactose using a GlcNAc-specific galactosyltransferase. BSA, bovine serum albumin.
FIGURE 2.
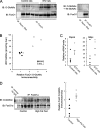
_O-_GlcNAcylation of FoxO1 is elevated in diabetes. A, FoxO1 was immunoprecipitated from control and STZ-induced diabetic rat livers and blotted with anti-O-GlcNAc antibodies (CTD110.6). Specificity was confirmed by GlcNAc competition. B, UDP-GlcNAc levels were determined from control and STZ-induced diabetic rat livers using capillary electrophoresis (y axis) and plotted against the densitometry analysis from C (x axis). C, RT-PCR analysis of_Pepck_ and G6pc expression show an elevated mRNA expression following STZ-induced diabetes. The dash represents the mean.D, FoxO1 was immunoprecipitated from control and high fat-fed mice and blotted with anti-O-GlcNAc antibodies (CTD110.6). Densitometry analysis reveals elevated GlcNAcylation of FoxO1. IB, immunoblotting.
FIGURE 3.
Insulin reduces FoxO1 GlcNAcylation elevated by high glucose. A, FLAG-FoxO1 was immunoprecipitated (IP) from Fao cells that were treated with insulin for 1 h. GlcNAcylation was detected by Western blotting using the CTD110.6 antibody. Glucose increases, whereas insulin decreases, FoxO1 GlcNAcylation. B, insulin treatment does not reduce total cellular GlcNAcylation in Fao cells. C, insulin treatment does not alter _O-_GlcNAcase activity in Fao cells. IB, immunoblotting.
FIGURE 4.
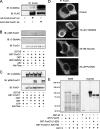
There is a complex interplay between phosphorylation and GlcNAcylation of FoxO1. A, the AKT-insensitive and constitutively nuclear FoxO1 mutant (T24A,S256A,S319A, called 3A) is hyper-GlcNAcylated in Fao cells.B, recombinant FoxO1 was in vitro GlcNAcylated then subsequently in vitro AKT labeled, subjected to SDS-PAGE, and blotted with anti-FoxO1 Ser(P)256, stripped, and then blotted with anti-O-GlcNAc antibodies (CTD110.6). Specificity of the Ser(P)256 antibody was confirmed by alkaline phosphatase treatment. C, recombinant FoxO1 was in vitro AKT-phosphorylated, then subsequently_in vitro O-_GlcNAc labeled, subjected to SDS-PAGE, and blotted with anti-O-GlcNAc antibodies (CTD110.6), stripped, and then blotted with anti-FoxO1 Ser(P)256 antibodies. Specificity of the Ser(P)256 antibody was confirmed by alkaline phosphatase treatment.In vitro AKT phosphorylation does not block in vitro OGT labeling of FoxO1. D, Fao cells were infected with adenovirus expressing FLAG-tagged FoxO1. 24 h after infection cells were serum-starved in RPMI + 0.5% bovine serum albumin for 4 h and then treated as indicated for an additional 16 h in RPMI + 0.5% bovine serum albumin. FoxO1 was visualized using anti-FoxO1 antibody. PUGNAc (an _O-_GlcNAcase inhibitor) or 25 m
m
glucose did not result increased nuclear translocation of FoxO1 under conditions tested. E, the FoxO3 isoform is an in vitro substrate for OGT. [3H]UDP-GlcNAc incorporation was detected by autoradiography following Coomassie G250 staining. The truncated (amino acids 1–525) form incorporated significantly less label, indicating a significant number of sites are in the C-terminal region. The mutant FoxO3 lacking AKT phosphorylation sites (T24A,S215A,S316A, or “triple mutant” or TM) incorporates [3H]UDP-GlcNAc similar to the truncated form, indicating that AKT phosphorylation sites are not significantly _in vitro O-_GlcNAc labeled. IB, immunoblotting;wt, wild type.
FIGURE 5.
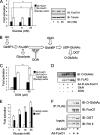
_O-_GlcNAc increases FoxO-dependent luciferase reporter transcription in HEK293 cells via the HBP. A, luciferase reporter activity is increased by high glucose for both wild type (wt) and 3A FoxO1 (all panels are plotted as relative luciferase activity normalized to β-galactosidase activity; error bars indicate standard errors; *, p < 0.05 by Student's t test). FoxO1 protein levels are unaffected. B, schematic of the UDP-GlcNAc synthesis pathway in which 2–5% of glucose that enters the cells is used for production of the donor sugar nucleotide. C, addition of 50 μ
m
DON, which inhibits the rate-limiting enzyme in UDP-GlcNAc synthesis (glutamine:fructose-6-phosphate amidotransferase, GFAT), reduces high glucose activation of luciferase activity. 10 m
m
glucosamine (GlcN) rescues activation of FoxO1. _D, O-_GlcNAc levels on FLAG-FoxO1 in Fao cells following treatment with 10 m
m
glucosamine and 50 μ
m
DON. E, overexpression of OGT increases FoxO-dependent luciferase activity. F, overexpression of OGT increases FoxO GlcNAcylation. CTD110.6 antibodies were used to detect FLAG-precipitated FoxO1. IP, immunoprecipitation; IB, immunoblotting.
FIGURE 6.
Glucose activates expression of FoxO1 target genes in an OGT-dependent manner. A, RT-PCR analysis shows gluconeogenic enzyme expression (G6pc and Pepck) is elevated by glucose in Fao hepatoma cells (all panels are relative expression normalized to 18 S rRNA; the_error bars_ indicate standard errors; *, p < 0.05 and **,p < 0.005 by Student's t test). B, RT-PCR analysis shows reactive oxygen species detoxification enzyme expression (Catalase, MnSOD) is elevated by glucose. Infection with an adenovirus expressing a dominant negative FoxO1 (Δ256) prevents activation of target genes by high glucose. C, RT-PCR analysis in OGT–/flox MEFs shows that OGT is required for high glucose activation of Catalase and MnSOD. D, RT-PCR analysis in Fao cells following OGT overexpression shows elevated Pepck and G6pc. E, RT-PCR analysis of mouse livers 7 days post-injection of adeno-OGT or GFP viruses.F, electrophoretic mobility shift assay with recombinant GlcNAcylated or naked GST-FoxO1 and a 300-base pair fragment of the IRE-luciferase reporter plasmid (used in Fig. 5) and visualized with ethidum bromide. G, electrophoretic mobility shift assay as in F with either 2 μg of GST + 6 μg of OGT, 2 μg of FoxO1, or 2 μg of FoxO1 + 6 μg of OGT.
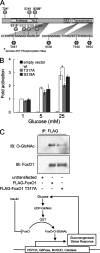
FIGURE 7.
FoxO1 is GlcNAcylated at multiple sites. A, schematic indicating four _O-_GlcNAc sites mapped using ETD MS/MS on human FoxO1 (Thr317, Ser550, Thr648, and Ser656). B, luciferase reporter assays of point mutants of FoxO1, indicating that mutation of Thr317 to alanine reduces activation by 25 m
m
glucose (expressed as relative activity normalized to β-galactosidase; error bars represent standard errors; *, p < 0.05 by Student's t test). C, HEK293 cells were transfected with FLAG-FoxO1 vectors containing the indicated site mutations, subjected to SDS-PAGE, and blotted using anti-O-GlcNAc antibodies (CTD110.6). IP, immunoprecipitation; IB, immunoblotting.
Similar articles
- A PGC-1alpha-O-GlcNAc transferase complex regulates FoxO transcription factor activity in response to glucose.
Housley MP, Udeshi ND, Rodgers JT, Shabanowitz J, Puigserver P, Hunt DF, Hart GW. Housley MP, et al. J Biol Chem. 2009 Feb 20;284(8):5148-57. doi: 10.1074/jbc.M808890200. Epub 2008 Dec 22. J Biol Chem. 2009. PMID: 19103600 Free PMC article. - O-GlcNAc protein modification in C2C12 myoblasts exposed to oxidative stress indicates parallels with endogenous antioxidant defense.
Peternelj TT, Marsh SA, Morais C, Small DM, Dalbo VJ, Tucker PS, Coombes JS. Peternelj TT, et al. Biochem Cell Biol. 2015 Feb;93(1):63-73. doi: 10.1139/bcb-2014-0106. Epub 2014 Oct 8. Biochem Cell Biol. 2015. PMID: 25453190 - FoxO1 deacetylation regulates thyroid hormone-induced transcription of key hepatic gluconeogenic genes.
Singh BK, Sinha RA, Zhou J, Xie SY, You SH, Gauthier K, Yen PM. Singh BK, et al. J Biol Chem. 2013 Oct 18;288(42):30365-30372. doi: 10.1074/jbc.M113.504845. Epub 2013 Aug 30. J Biol Chem. 2013. PMID: 23995837 Free PMC article. - O-GlcNAc modification of FoxO1 increases its transcriptional activity: a role in the glucotoxicity phenomenon?
Kuo M, Zilberfarb V, Gangneux N, Christeff N, Issad T. Kuo M, et al. Biochimie. 2008 May;90(5):679-85. doi: 10.1016/j.biochi.2008.03.005. Epub 2008 Mar 21. Biochimie. 2008. PMID: 18359296 Review. - Redox regulation of FoxO transcription factors.
Klotz LO, Sánchez-Ramos C, Prieto-Arroyo I, Urbánek P, Steinbrenner H, Monsalve M. Klotz LO, et al. Redox Biol. 2015 Dec;6:51-72. doi: 10.1016/j.redox.2015.06.019. Epub 2015 Jul 3. Redox Biol. 2015. PMID: 26184557 Free PMC article. Review.
Cited by
- Protein Substrates Engage the Lumen of O-GlcNAc Transferase's Tetratricopeptide Repeat Domain in Different Ways.
Joiner CM, Hammel FA, Janetzko J, Walker S. Joiner CM, et al. Biochemistry. 2021 Mar 23;60(11):847-853. doi: 10.1021/acs.biochem.0c00981. Epub 2021 Mar 12. Biochemistry. 2021. PMID: 33709700 Free PMC article. - The role of liver fructose-1,6-bisphosphatase in regulating appetite and adiposity.
Visinoni S, Khalid NF, Joannides CN, Shulkes A, Yim M, Whitehead J, Tiganis T, Lamont BJ, Favaloro JM, Proietto J, Andrikopoulos S, Fam BC. Visinoni S, et al. Diabetes. 2012 May;61(5):1122-32. doi: 10.2337/db11-1511. Diabetes. 2012. PMID: 22517657 Free PMC article. - The Beginner's Guide to _O_-GlcNAc: From Nutrient Sensitive Pathway Regulation to Its Impact on the Immune System.
Mannino MP, Hart GW. Mannino MP, et al. Front Immunol. 2022 Jan 31;13:828648. doi: 10.3389/fimmu.2022.828648. eCollection 2022. Front Immunol. 2022. PMID: 35173739 Free PMC article. Review. - Protein O-linked β-N-acetylglucosamine: a novel effector of cardiomyocyte metabolism and function.
Darley-Usmar VM, Ball LE, Chatham JC. Darley-Usmar VM, et al. J Mol Cell Cardiol. 2012 Mar;52(3):538-49. doi: 10.1016/j.yjmcc.2011.08.009. Epub 2011 Aug 22. J Mol Cell Cardiol. 2012. PMID: 21878340 Free PMC article. Review. - Site-specific GlcNAcylation of human erythrocyte proteins: potential biomarker(s) for diabetes.
Wang Z, Park K, Comer F, Hsieh-Wilson LC, Saudek CD, Hart GW. Wang Z, et al. Diabetes. 2009 Feb;58(2):309-17. doi: 10.2337/db08-0994. Epub 2008 Nov 4. Diabetes. 2009. PMID: 18984734 Free PMC article.
References
- Zhang, W., Patil, S., Chauhan, B., Guo, S., Powell, D. R., Le, J., Klotsas, A., Matika, R., Xiao, X., Franks, R., Heidenreich, K. A., Sajan, M. P., Farese, R. V., Stolz, D. B., Tso, P., Koo, S. H., Montminy, M., and Unterman, T. G. (2006) J. Biol. Chem. 281 10105–10117 - PubMed
- Puig, O., and Tjian, R. (2006) Cell Cycle 5 503–505 - PubMed
- Lehtinen, M. K., Yuan, Z., Boag, P. R., Yang, Y., Villen, J., Becker, E. B., DiBacco, S., de la Iglesia, N., Gygi, S., Blackwell, T. K., and Bonni, A. (2006) Cell 125 987–1001 - PubMed
- Tothova, Z., Kollipara, R., Huntly, B. J., Lee, B. H., Castrillon, D. H., Cullen, D. E., McDowell, E. P., Lazo-Kallanian, S., Williams, I. R., Sears, C., Armstrong, S. A., Passegue, E., DePinho, R. A., and Gilliland, D. G. (2007) Cell 128 325–339 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- DK61671/DK/NIDDK NIH HHS/United States
- GM37537/GM/NIGMS NIH HHS/United States
- R37 HD013563/HD/NICHD NIH HHS/United States
- HD13563/HD/NICHD NIH HHS/United States
- R01 GM037537/GM/NIGMS NIH HHS/United States
- R01 DK061671/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous