TRIM21 is an IgG receptor that is structurally, thermodynamically, and kinetically conserved - PubMed (original) (raw)
Comparative Study
. 2008 Apr 22;105(16):6045-50.
doi: 10.1073/pnas.0800159105. Epub 2008 Apr 17.
Affiliations
- PMID: 18420815
- PMCID: PMC2329685
- DOI: 10.1073/pnas.0800159105
Comparative Study
TRIM21 is an IgG receptor that is structurally, thermodynamically, and kinetically conserved
Anthony H Keeble et al. Proc Natl Acad Sci U S A. 2008.
Abstract
The newly identified tripartite motif (TRIM) family of proteins mediate innate immunity and other critical cellular functions. Here we show that TRIM21, which mediates the autoimmune diseases rheumatoid arthritis, systemic lupus erythematosus, and Sjögren's syndrome, is a previously undescribed IgG receptor with a binding mechanism unlike known mammalian Fcgamma receptors. TRIM21 simultaneously targets conserved hot-spot residues on both Ig domains of the Fc fragment using a PRYSPRY domain with a preformed multisite interface. The binding sites on both TRIM21 and Fc are highly conserved to the extent that the proteins are functionally interchangeable through murine, canine, primate, and human species. Pre-steady-state analysis exposes mechanistic conservation at the level of individual residues, which make the same energetic and kinetic contributions to binding despite varying in sequence. Together, our results reveal that TRIM21 is a previously undescribed type of IgG receptor based on a non-Ig scaffold whose interaction at the fundamental level-structural, thermodynamic, and kinetic-is evolutionarily conserved.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
Comparison of free and bound mouse TRIM21 structures. (Left) Superposition of free (wheat) with bound (orange) PRYSPRY domain. Binding-site loops and residues making important contributions are annotated. (Right) Superposition of TRIM21:Fc complex (orange and red, respectively) with one Fc heavy chain from mouse IgG structure 1IGT (24) (pink). A superposition of the free structure is shown in yellow.
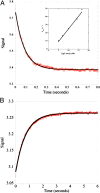
Fig. 2.
Pre-steady-state kinetics of mouse TRIM21:Fc interaction. (A) Stopped-flow spectrofluorimetry was used to measure the rapid fluorescence quench upon mouse TRIM21–mouse IgG association (see Materials and Methods for details). The data (red circles) fit to a single exponential (solid black line). Linear regression of the linear increase in _k_obs (Inset) yields an association rate constant (_k_on) of 3.16 × 106 M−1s−1. (B) Measurement of the dissociation rate constant (_k_off) for mouse TRIM21–mouse IgG binding in a stopped-flow experiment by competition with excess protein A. This yields a _k_off of 1.35 s−1 resulting in a kinetic _K_d of 437 nM, close to that observed by ITC.
Fig. 3.
Hot-spot, electrostatic potential, and hydrophobicity conservation in the TRIM21 PRYSPRY binding site. Human (Left) is compared with mouse (Right). The molecular surface of the binding site is shown and colored on a red scale for ΔΔG, on a green scale for hydrophobicity, and on a red to blue (negative to positive) scale for electrostatic potential.
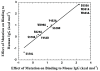
Fig. 4.
Effect of site-directed mutagenesis on cognate versus noncognate species TRIM21:IgG interaction. When the effects of mouse TRIM21 mutations on binding mouse IgG (cognate; x axis) are plotted against the effect on binding human IgG (noncognate; y axis) there exists a striking linear correlation (_R_2 = 0.98) suggesting that the same interactions are used to stabilize both complexes.
Fig. 5.
Sequence alignment of mouse and human TRIM21. Identical residues are shaded gray. The positions of the variable binding loops are shaded purple.
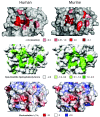
Fig. 6.
Comparison of mouse and human TRIM21. (Left) Superposition of mouse (orange) and human (yellow) PRYSPRY domains. Where the sequences diverge the annotations are formatted so that the first residue is for human and the second is for mouse; e.g., D/S326 indicates an aspartic acid in human and a serine in mouse. (Right) Superposition of mouse (red and orange) and human (pink and yellow) complexes with Fc.
Similar articles
- Structural basis for PRYSPRY-mediated tripartite motif (TRIM) protein function.
James LC, Keeble AH, Khan Z, Rhodes DA, Trowsdale J. James LC, et al. Proc Natl Acad Sci U S A. 2007 Apr 10;104(15):6200-5. doi: 10.1073/pnas.0609174104. Epub 2007 Mar 30. Proc Natl Acad Sci U S A. 2007. PMID: 17400754 Free PMC article. - TRIM21 is a trimeric protein that binds IgG Fc via the B30.2 domain.
Rhodes DA, Trowsdale J. Rhodes DA, et al. Mol Immunol. 2007 Mar;44(9):2406-14. doi: 10.1016/j.molimm.2006.10.013. Epub 2006 Nov 21. Mol Immunol. 2007. PMID: 17118455 - The Sjögren's syndrome-associated autoantigen Ro52/TRIM21 modulates follicular B cell homeostasis and immunoglobulin production.
Brauner S, Ivanchenko M, Thorlacius GE, Ambrosi A, Wahren-Herlenius M. Brauner S, et al. Clin Exp Immunol. 2018 Dec;194(3):315-326. doi: 10.1111/cei.13211. Epub 2018 Oct 16. Clin Exp Immunol. 2018. PMID: 30178506 Free PMC article. - TRIM21: a cytosolic Fc receptor with broad antibody isotype specificity.
Foss S, Watkinson R, Sandlie I, James LC, Andersen JT. Foss S, et al. Immunol Rev. 2015 Nov;268(1):328-39. doi: 10.1111/imr.12363. Immunol Rev. 2015. PMID: 26497531 Free PMC article. Review. - Structural analysis of Fc/FcγR complexes: a blueprint for antibody design.
Caaveiro JM, Kiyoshi M, Tsumoto K. Caaveiro JM, et al. Immunol Rev. 2015 Nov;268(1):201-21. doi: 10.1111/imr.12365. Immunol Rev. 2015. PMID: 26497522 Review.
Cited by
- Structure of the rhesus monkey TRIM5α PRYSPRY domain, the HIV capsid recognition module.
Biris N, Yang Y, Taylor AB, Tomashevski A, Guo M, Hart PJ, Diaz-Griffero F, Ivanov DN. Biris N, et al. Proc Natl Acad Sci U S A. 2012 Aug 14;109(33):13278-83. doi: 10.1073/pnas.1203536109. Epub 2012 Jul 30. Proc Natl Acad Sci U S A. 2012. PMID: 22847415 Free PMC article. - Crystal structure of TRIM20 C-terminal coiled-coil/B30.2 fragment: implications for the recognition of higher order oligomers.
Weinert C, Morger D, Djekic A, Grütter MG, Mittl PR. Weinert C, et al. Sci Rep. 2015 Jun 4;5:10819. doi: 10.1038/srep10819. Sci Rep. 2015. PMID: 26043233 Free PMC article. - Regulation of virus neutralization and the persistent fraction by TRIM21.
McEwan WA, Hauler F, Williams CR, Bidgood SR, Mallery DL, Crowther RA, James LC. McEwan WA, et al. J Virol. 2012 Aug;86(16):8482-91. doi: 10.1128/JVI.00728-12. Epub 2012 May 30. J Virol. 2012. PMID: 22647693 Free PMC article. - Molecular basis of antibody mediated immunity against Ehrlichia chaffeensis involves species-specific linear epitopes in tandem repeat proteins.
Kuriakose JA, Zhang X, Luo T, McBride JW. Kuriakose JA, et al. Microbes Infect. 2012 Oct;14(12):1054-63. doi: 10.1016/j.micinf.2012.05.012. Epub 2012 May 29. Microbes Infect. 2012. PMID: 22658957 Free PMC article. - Optophysiology: Illuminating cell physiology with optogenetics.
Tan P, He L, Huang Y, Zhou Y. Tan P, et al. Physiol Rev. 2022 Jul 1;102(3):1263-1325. doi: 10.1152/physrev.00021.2021. Epub 2022 Jan 24. Physiol Rev. 2022. PMID: 35072525 Free PMC article. Review.
References
- Nisole S, Stoye JP, Saib A. TRIM family proteins: Retroviral restriction and antiviral defence. Nat Rev Microbiol. 2005;3:799–808. - PubMed
- Meroni G, Diez-Roux G. TRIM/RBCC, a novel class of ‘single protein RING finger’ E3 ubiquitin ligases. BioEssays. 2005;27:1147–1157. - PubMed
- Stremlau M, et al. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 2004;427:848–853. - PubMed
- Trockenbacher A, et al. MID1, mutated in Opitz syndrome, encodes an ubiquitin ligase that targets phosphatase 2A for degradation. Nat Genet. 2001;29:287–294. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases