Identification of a bacterial-like HslVU protease in the mitochondria of Trypanosoma brucei and its role in mitochondrial DNA replication - PubMed (original) (raw)
Identification of a bacterial-like HslVU protease in the mitochondria of Trypanosoma brucei and its role in mitochondrial DNA replication
Ziyin Li et al. PLoS Pathog. 2008.
Abstract
ATP-dependent protease complexes are present in all living organisms, including the 26S proteasome in eukaryotes, Archaea, and Actinomycetales, and the HslVU protease in eubacteria. The structure of HslVU protease resembles that of the 26S proteasome, and the simultaneous presence of both proteases in one organism was deemed unlikely. However, HslVU homologs have been identified recently in some primordial eukaryotes, though their potential function remains elusive. We characterized the HslVU homolog from Trypanosoma brucei, a eukaryotic protozoan parasite and the causative agent of human sleeping sickness. TbHslVU has ATP-dependent peptidase activity and, like its bacterial counterpart, has essential lysine and N-terminal threonines in the catalytic subunit. By epitope tagging, TbHslVU localizes to mitochondria and is associated with the mitochondrial genome, kinetoplast DNA (kDNA). RNAi of TbHslVU dramatically affects the kDNA by causing over-replication of the minicircle DNA. This leads to defects in kDNA segregation and, subsequently, to continuous network growth to an enormous size. Multiple discrete foci of nicked/gapped minicircles are formed on the periphery of kDNA disc, suggesting a failure in repairing the gaps in the minicircles for kDNA segregation. TbHslVU is a eubacterial protease identified in the mitochondria of a eukaryote. It has a novel function in regulating mitochondrial DNA replication that has never been observed in other organisms.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
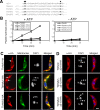
Figure 1. Enzymatic activity and intracellular localization of TbHslVU.
(A). TbHslV contains the conserved threonine and lysine residues (arrows) found essential for the activities of HslV in E. coli and the β-subunits of 20S CP in T. brucei. (B). The ATP-dependent peptidase activity of TbHslV. Wild type and three TbHslV mutants T20A, T21A and K53A were expressed as HA-tagged proteins in T. brucei, immunoprecipitated and assayed for hydrolysis of Cbz-Gly-Gly-Leu-AMC. (C). Cells stably expressing TbHslVU-HA were labeled for mitochondria with Mitotracker green dye (green), immunostained with anti-HA mAb for HA-tagged proteins (red) and counterstained with DAPI for DNA (blue). Arrows indicate the focal points of HA-staining corresponding to the positions of kinetoplasts (arrowheads). (D). Cells expressing TbHslVU-HA with the putative mitochondrial targeting sequences deleted were stained with anti-HA antibody (red) and counterstained with DAPI. Bars: 2 µm.
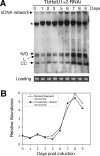
Figure 2. RNAi knockdown of TbHslVU affects cell growth and kinetoplast morphology.
(A). Cells were grown in the presence (+) of tetracycline to induce RNAi for 7 days, and cell growth was monitored daily. Northern blots were performed to assess levels of TbHslV, TbHslU1 and TbHslU2 mRNA before (−) and after (+) 2 days of RNAi (insets). (B–E). Un-induced control cells (B) and cells after RNAi induction for 7 days (C) were labeled with YL1/2 antibody for basal bodies (BB, arrowheads) and counterstained with DAPI for nucleus (N) and kinetoplast (arrows). (D). Two TbHslVU RNAi cells at the final stage of cell division were still connected by a thin thread of kinetoplast DNA (arrows) between two basal bodies (arrowheads) in two well-separated cells. Bars: 2 µm. (E). Tabulation of RNAi cells with kinetoplasts in varying sizes and morphologies. Approximately 200 cells were counted at each time point and the data represent averages from three independent experiments.
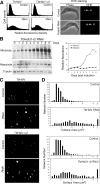
Figure 3. TbHslVU RNAi led to heterogeneously sized kinetoplasts.
(A). Flow cytometry analysis of DHE stained cells. A total of 25,000 cells were counted in each experiment (Left panel). DHE stains exclusively the kinetoplasts in control and RNAi cells (Right panel). (B). Southern analysis of changes in minicircle and maxicircle DNA content during TbHslU RNAi. The kinetics of minicircle (open circle) and maxicircle (filled square) accumulation are presented to the right of the Southern blots. (C). DAPI staining of the isolated kDNA networks. (D). Surface areas of the isolated kDNA networks stained with DAPI, and measured with the NIH Image software.
Figure 4. Electron microscopic examination of kDNA networks from the control and TbHslV RNAi cells.
Methods were described by . (A, B, E, F) kDNA networks from cells after 7 days of TbHslV RNAi. (H) A kDNA network from an un-induced control cell. (C, D) Enlargements of the network in A, corresponding to the areas outlined in white framed boxes. (G) Enlargement of the network in F framed in a white box. The arrow in G indicates a maxicircle. Scale bars for A, B, E, F, and H, 2 µm and for C, D, and G, 0.5 µm.
Figure 5. Effect of TbHslU1+2 double RNAi on free minicircle replication intermediates.
(A). Total DNA was fractionated on an agarose/ethidium gel and a Southern blot was probed for minicircles. N/G, nicked/gapped minicircles; L, linearized minicircles; CC, covalently-closed minicircles, *, nonspecific hybridization to nuclear DNA. The nicked/gapped minicircles form a doublet with the lower component possibly linearized minicircles. Since it is present prior to RNAi induction (day 0), it is likely unrelated to RNAi. (B). Quantitation (by Phosphorimager) of bands from A. The nicked/gapped species includes both components of the doublet.
Figure 6. In situ TdT-catalyzed Fluorescein-dUTP labeling in cells after 7 days of TbHslV RNAi.
(A). The control cells (a–f). (B, C). The TbHslV RNAi cells (g–p). Percentages of TdT-labeled cells in control and TbHslV RNAi cells are presented (k). Bars: 2 µm.
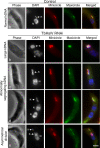
Figure 7. In situ detection of minicircles and maxicircles in the kinetoplast by FISH.
Cells were fixed, probed for minicircles (red) or maxicircles (green), and counterstained with DAPI. kDNA is indicated with a K and nucleus with an N. Bar: 2 µm. FISH does not detect covalently-closed DNA minicircles because they are non-denaturable . This is likely also true for maxicircles. Thus the FISH signal may not be proportional to the total populations of minicircles and maxicircles. Bars: 2 µm.
Similar articles
- Characterization of the novel mitochondrial genome replication factor MiRF172 in Trypanosoma brucei.
Amodeo S, Jakob M, Ochsenreiter T. Amodeo S, et al. J Cell Sci. 2018 Apr 25;131(8):jcs211730. doi: 10.1242/jcs.211730. J Cell Sci. 2018. PMID: 29626111 Free PMC article. - Trypanosoma brucei Tim50 Possesses PAP Activity and Plays a Critical Role in Cell Cycle Regulation and Parasite Infectivity.
Tripathi A, Singha UK, Cooley A, Gillyard T, Krystofiak E, Pratap S, Davis J, Chaudhuri M. Tripathi A, et al. mBio. 2021 Oct 26;12(5):e0159221. doi: 10.1128/mBio.01592-21. Epub 2021 Sep 14. mBio. 2021. PMID: 34517757 Free PMC article. - The bacterial-like HslVU protease complex subunits are involved in the control of different cell cycle events in trypanosomatids.
Mbang-Benet DE, Sterkers Y, Morelle C, Kebe NM, Crobu L, Portalès P, Coux O, Hernandez JF, Meghamla S, Pagès M, Bastien P. Mbang-Benet DE, et al. Acta Trop. 2014 Mar;131:22-31. doi: 10.1016/j.actatropica.2013.11.017. Epub 2013 Dec 1. Acta Trop. 2014. PMID: 24299926 - Mitochondrial genome maintenance-the kinetoplast story.
Amodeo S, Bregy I, Ochsenreiter T. Amodeo S, et al. FEMS Microbiol Rev. 2023 Nov 1;47(6):fuac047. doi: 10.1093/femsre/fuac047. FEMS Microbiol Rev. 2023. PMID: 36449697 Free PMC article. Review. - Malleable mitochondrion of Trypanosoma brucei.
Verner Z, Basu S, Benz C, Dixit S, Dobáková E, Faktorová D, Hashimi H, Horáková E, Huang Z, Paris Z, Peña-Diaz P, Ridlon L, Týč J, Wildridge D, Zíková A, Lukeš J. Verner Z, et al. Int Rev Cell Mol Biol. 2015;315:73-151. doi: 10.1016/bs.ircmb.2014.11.001. Epub 2015 Feb 7. Int Rev Cell Mol Biol. 2015. PMID: 25708462 Review.
Cited by
- Trypanosoma brucei Tb927.2.6100 is an essential protein associated with kinetoplast DNA.
Beck K, Acestor N, Schulfer A, Anupama A, Carnes J, Panigrahi AK, Stuart K. Beck K, et al. Eukaryot Cell. 2013 Jul;12(7):970-8. doi: 10.1128/EC.00352-12. Epub 2013 May 6. Eukaryot Cell. 2013. PMID: 23650088 Free PMC article. - Trypanosomes have six mitochondrial DNA helicases with one controlling kinetoplast maxicircle replication.
Liu B, Wang J, Yaffe N, Lindsay ME, Zhao Z, Zick A, Shlomai J, Englund PT. Liu B, et al. Mol Cell. 2009 Aug 28;35(4):490-501. doi: 10.1016/j.molcel.2009.07.004. Epub 2009 Jul 30. Mol Cell. 2009. PMID: 19646907 Free PMC article. - Unwinding the functions of the Pif1 family helicases.
Bochman ML, Sabouri N, Zakian VA. Bochman ML, et al. DNA Repair (Amst). 2010 Mar 2;9(3):237-49. doi: 10.1016/j.dnarep.2010.01.008. Epub 2010 Jan 25. DNA Repair (Amst). 2010. PMID: 20097624 Free PMC article. Review. - Structural and biochemical analyses of the eukaryotic heat shock locus V (HslV) from Trypanosoma brucei.
Sung KH, Lee SY, Song HK. Sung KH, et al. J Biol Chem. 2013 Aug 9;288(32):23234-43. doi: 10.1074/jbc.M113.484832. Epub 2013 Jul 1. J Biol Chem. 2013. PMID: 23818520 Free PMC article. - Unexpected Evolution of Lesion-Recognition Modules in Eukaryotic NER and Kinetoplast DNA Dynamics Proteins from Bacterial Mobile Elements.
Krishnan A, Burroughs AM, Iyer LM, Aravind L. Krishnan A, et al. iScience. 2018 Nov 30;9:192-208. doi: 10.1016/j.isci.2018.10.017. Epub 2018 Oct 23. iScience. 2018. PMID: 30396152 Free PMC article.
References
- Coux O, Tanaka K, Goldberg AL. Structure and functions of the 20S and 26S proteasomes. Annu Rev Biochem. 1996;65:801–847. - PubMed
- De Mot R, Nagy I, Walz J, Baumeister W. Proteasomes and other self-compartmentalizing proteases in prokaryotes. Trends Microbiol. 1999;7:88–92. - PubMed
- Voges D, Zwickl P, Baumeister W. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu Rev Biochem. 1999;68:1015–1068. - PubMed
- Bochtler M, Hartmann C, Song HK, Bourenkov GP, Bartunik HD, et al. The structures of HsIU and the ATP-dependent protease HsIU-HsIV. Nature. 2000;403:800–805. - PubMed
- Bochtler M, Ditzel L, Groll M, Hartmann C, Huber R. The proteasome. Annu Rev Biophys Biomol Struct. 1999;28:295–317. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI058613/AI/NIAID NIH HHS/United States
- R01 AI021786/AI/NIAID NIH HHS/United States
- R01 AI-021786/AI/NIAID NIH HHS/United States
- R01 AI-058613/AI/NIAID NIH HHS/United States
- R56 AI021786/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases