The Caenorhabditis elegans rsd-2 and rsd-6 genes are required for chromosome functions during exposure to unfavorable environments - PubMed (original) (raw)
The Caenorhabditis elegans rsd-2 and rsd-6 genes are required for chromosome functions during exposure to unfavorable environments
Wang Han et al. Genetics. 2008 Apr.
Abstract
In Caenorhabditis elegans, exogenous dsRNA can elicit systemic RNAi, a process that requires the function of many genes. Considering that the activities of many of these genes are also required for normal development, it is surprising that exposure to high concentrations of dsRNA does not elicit adverse consequences to animals. Here, we report inducible phenotypes in attenuated C. elegans strains reared in environments that include nonspecific dsRNA and elevated temperature. Under these conditions, chromosome integrity is compromised in RNAi-defective strains harboring mutations in rsd-2 or rsd-6. Specifically, rsd-2 mutants display defects in transposon silencing, while meiotic chromosome disjunction is affected in rsd-6 mutants. RSD-2 proteins localize to multiple cellular compartments, including the nucleolus and cytoplasmic compartments that, in part, are congruent with calreticulin and HAF-6. We considered that the RNAi defects in rsd-2 mutants might have relevance to membrane-associated functions; however, endomembrane compartmentalization and endocytosis/exocytosis markers in rsd-2 and rsd-6 mutants appear normal. The mutants also possess environmentally sensitive defects in cell-autonomous RNAi elicited from transgene-delivered dsRNAs. Thus, the ultimate functions of rsd-2 and rsd-6 in systemic RNAi are remarkably complex and environmentally responsive.
Figures
Figure 1.—
Novel mutations in sid-1 and rsd-2 and identification of two mRNA isoforms for rsd-2. (A) The relative positions of the mutations are indicated in this diagram of intron/exon configurations for sid-1 and rsd-2 genes (not to scale). (Bottom) The relative positions of primers (arrows) are indicated above the diagram; SL1 splice acceptor regions are indicated below the diagram. (B) 5′-end mapping of rsd-2 mRNA. Experiments were performed following RT–PCR protocols 1–3, respectively (see
materials and methods
). The primers utilized in each final PCR reaction are indicated. Asterisks highlight the positions expected for long- and short-form cDNAs. Expected sizes of bands: SL1/281 (lane 1)—long-form cDNA, 1068 bp; short-form cDNA, 510 bp (the primers will amplify both forms); SL1 or SL2/279 (lanes 3–6)—long-form cDNA, 848 bp; short-form cDNA, 294 bp (the primers will amplify both forms); 581/583 (lane 7)—long-form cDNA, 4085 bp (only one form will amplify); 582/583 (lane 8)—long- and short-form cDNA, 3585 bp (both forms will amplify to produce products with identical sequence). All PCR primers flank introns; therefore, genomic and cDNA fragments can clearly be distinguished by size.
Figure 2.—
Transgene rescue and GFP reporters. (A) Configuration of sequences used in transgenic strains. The let-858 promoter drives ubiquitous expression (K
elly
et al. 1997). GFP reporter constructs were generated for both protein isoforms, with GFP sequences fused at the N or C terminus. (B) An example of transgene rescue of RNAi defects in rsd-2 mutants assessed using pop-1 food. Wild-type animals displayed sterility in response to ingestion of pop-1 food, while rsd-2(ne319) mutants were RNAi defective and produced progeny. rsd-2(ne319) mutants harboring a transgene capable of expressing both RSD-2A and RSD-2B proteins displayed RNAi. Similar results are observed when the experiment is performed at 20° (shown) vs. 25°.
Figure 3.—
Residual RNAi activity in mutants and dosage sensitivities. pop-1 dsRNA was introduced into animals using injection, soaking, and feeding methodologies to better assess the nature of the RNAi defects in mutants. (A) A dilution series of pop-1 dsRNA was prepared and injected into the intestine of animals with the indicated genotype. Each injected animal was placed onto a standard culture plate, and the ensuing progeny were counted. Bars represent the average number of progeny produced per experiment. An average of 11 animals was injected in each experiment (6–19 animals). Error bars denote the SD. (B) Animals of the indicated genotypes were soaked in 200 ng/μl of pop-1 dsRNA and incubated overnight at 15°. Each treated animal was placed onto a separate culture plate, and the progeny were tabulated. Bars represent the average number of progeny from two separate experiments. An average of 16 treated animals were analyzed (11–21 per experiment). Error bars denote SD. (C) Three or four L1-stage animals were placed on pop-1 food and reared at 20°. The average number of progeny produced per treated adult was tabulated. Bars represent an average of three independent assessments; error bars denote the SD.
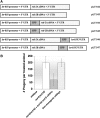
Figure 4.—
Conditional phenotypes in rsd-2 and rsd-6 mutants. (A) Mutants reared on unc-22 food display RNAi defects that are temperature sensitive in nature. Animals were placed as L1 larvae on unc-22 food and were reared at the indicated temperatures. The percentage of progeny animals displaying a twitching phenotype was scored. Bars represent the average of two experiments; n = 80–257 animals; error bars denote SD. (B) Temperature- and dsRNA-sensitive nondisjunction phenotypes in mutants. Strains of the indicated genotype were reared at 20° or 25° on normal culture plates or on plates seeded with bacteria designed to overexpress nonspecific dsRNA corresponding in sequence to a bacterial tetracycline resistance gene. The culture plates were checked each day for males, which were counted and immediately removed; n = 300–3300. Bars represent the average of three experiments with SD. (C) Temperature-sensitive mutator activity in rsd-2 mutants. All animals harbored the unc-22(st136) transposon-insertion mutation. Animals were cultured on standard plates as described and the percentages of revertant animal were scored. (D) Temperature-sensitive sterility is observed in rsd-6, but not in rsd-2 mutants. Bars represent the average number of F1 progeny produced from adults reared on standard culture plates at the indicated temperature. An average of three experiments is represented; error bars denote the range. Brood sizes were normalized to the average wild-type value at each temperature.
Figure 4.—
Conditional phenotypes in rsd-2 and rsd-6 mutants. (A) Mutants reared on unc-22 food display RNAi defects that are temperature sensitive in nature. Animals were placed as L1 larvae on unc-22 food and were reared at the indicated temperatures. The percentage of progeny animals displaying a twitching phenotype was scored. Bars represent the average of two experiments; n = 80–257 animals; error bars denote SD. (B) Temperature- and dsRNA-sensitive nondisjunction phenotypes in mutants. Strains of the indicated genotype were reared at 20° or 25° on normal culture plates or on plates seeded with bacteria designed to overexpress nonspecific dsRNA corresponding in sequence to a bacterial tetracycline resistance gene. The culture plates were checked each day for males, which were counted and immediately removed; n = 300–3300. Bars represent the average of three experiments with SD. (C) Temperature-sensitive mutator activity in rsd-2 mutants. All animals harbored the unc-22(st136) transposon-insertion mutation. Animals were cultured on standard plates as described and the percentages of revertant animal were scored. (D) Temperature-sensitive sterility is observed in rsd-6, but not in rsd-2 mutants. Bars represent the average number of F1 progeny produced from adults reared on standard culture plates at the indicated temperature. An average of three experiments is represented; error bars denote the range. Brood sizes were normalized to the average wild-type value at each temperature.
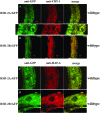
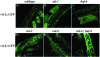
Figure 5.—
Subcellular localization of RSD-2 proteins. (A–D) Double-labeling experiments using anti-GFP antibodies and RSD-2∷GFP reporters. A second primary antibody targeted calreticulin (A and B) or HAF-6 (C and D). (A and B) Staining experiments in wild-type animals expressing a GFP-tagged version of RSD-2A. (C and D) Wild-type animals expressing RSD-2B∷GFP. The images were compiled in Photoshop. All images are from sections of adult, hermaphrodite intestine. (A, bottom) A higher magnification section from the top image. All transgenes used in these experiments provided RNAi activity when tested in an rsd-2(ne319) background (Table 1).
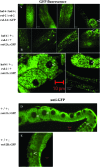
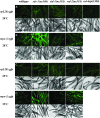
Figure 6.—
Subcellular localization of GFP-tagged RSD-2 protein in live animals. GFP fluorescence was observed in live animals harboring an integrated transgene expressing RSD-2A∷GFP fusion protein in all cells. (A) GFP fluorescence in the intestine and embryos (carets) of haf-6(ne335); rsd-2(ne319) double mutants. (B) Similar patterns are observed in the heterozygous progeny of animals in A mated to wild-type males. (C) Nucleolar GFP fluorescence was observed in some cells (left embryo), but not others (right oocyte) of live animals. Nucleolar staining is also observed in germline tissue (D) or intestine (E) of fixed animals harboring an extrachromosomal array expressing RSD-2∷GFP.
Figure 7.—
Vitellogenin localization patterns in wild-type and mutant animals. GFP fluorescence was observed in live animals harboring a transgene expressing a GFP-tagged version of vitellogenin. Intestinal tissue and mature oocytes or embryos are indicated by carets. GFP fluorescence was observed in intestine and oocytes/embryos for all genotypes: (A) wild type, (B) sid-1(qt2), (C) haf-6(ne335), (D) rsd-2(ne319), (E) rsd-6(pk3300), and (F) rsd-6(pk3300); haf-6(ne335) double mutant.
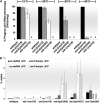
Figure 8.—
Assays for systemic and cell-autonomous RNAi. Populations of animals were photographed to allow for comparisons of GFP intensity. All animals harbor a transgene expressing gfp dsRNA in the muscle. The animals are homozygous for the alleles listed at the top of the figure. Systemic RNAi responses were assessed using an integrated transgene that expresses GFP in all cells (A and C); cell-autonomous RNAi was assessed using an integrated transgene that expresses GFP in muscle (B and D). Animals harboring both the rsd-6 mutation and the myo-3∷gfp hairpin array were not generated; the array is inserted close to the rsd-6 mutation and recombinants could not be generated. Animals were reared for several generations at either 20° (A and B) or 25° (C and D). The fluorescent images in A–D were captured and processed in Photoshop identically, as described in
materials and methods
. (E) Individual animals were visualized using fluorescent microscopy and scored as Bright (little RNAi activity), Medium (some RNAi activity), or Dim (virtually no RNAi activity) as described in
materials and methods
. Each strain was reared at 20° or 25°, as indicated, for several generations. n ≥ 50 for each bar. Bars represent the average of two experiments; error bars indicate SD.
Figure 8.—
Assays for systemic and cell-autonomous RNAi. Populations of animals were photographed to allow for comparisons of GFP intensity. All animals harbor a transgene expressing gfp dsRNA in the muscle. The animals are homozygous for the alleles listed at the top of the figure. Systemic RNAi responses were assessed using an integrated transgene that expresses GFP in all cells (A and C); cell-autonomous RNAi was assessed using an integrated transgene that expresses GFP in muscle (B and D). Animals harboring both the rsd-6 mutation and the myo-3∷gfp hairpin array were not generated; the array is inserted close to the rsd-6 mutation and recombinants could not be generated. Animals were reared for several generations at either 20° (A and B) or 25° (C and D). The fluorescent images in A–D were captured and processed in Photoshop identically, as described in
materials and methods
. (E) Individual animals were visualized using fluorescent microscopy and scored as Bright (little RNAi activity), Medium (some RNAi activity), or Dim (virtually no RNAi activity) as described in
materials and methods
. Each strain was reared at 20° or 25°, as indicated, for several generations. n ≥ 50 for each bar. Bars represent the average of two experiments; error bars indicate SD.
Similar articles
- Transgene-Assisted Genetic Screen Identifies rsd-6 and Novel Genes as Key Components of Antiviral RNA Interference in Caenorhabditis elegans.
Long T, Meng F, Lu R. Long T, et al. J Virol. 2018 Aug 16;92(17):e00416-18. doi: 10.1128/JVI.00416-18. Print 2018 Sep 1. J Virol. 2018. PMID: 29950414 Free PMC article. - Endomembrane-associated RSD-3 is important for RNAi induced by extracellular silencing RNA in both somatic and germ cells of Caenorhabditis elegans.
Imae R, Dejima K, Kage-Nakadai E, Arai H, Mitani S. Imae R, et al. Sci Rep. 2016 Jun 16;6:28198. doi: 10.1038/srep28198. Sci Rep. 2016. PMID: 27306325 Free PMC article. - The Caenorhabditis elegans RDE-10/RDE-11 complex regulates RNAi by promoting secondary siRNA amplification.
Zhang C, Montgomery TA, Fischer SE, Garcia SM, Riedel CG, Fahlgren N, Sullivan CM, Carrington JC, Ruvkun G. Zhang C, et al. Curr Biol. 2012 May 22;22(10):881-90. doi: 10.1016/j.cub.2012.04.011. Epub 2012 Apr 26. Curr Biol. 2012. PMID: 22542102 Free PMC article. - Systemic RNAi in Caenorhabditis elegans.
Hunter CP, Winston WM, Molodowitch C, Feinberg EH, Shih J, Sutherlin M, Wright AJ, Fitzgerald MC. Hunter CP, et al. Cold Spring Harb Symp Quant Biol. 2006;71:95-100. doi: 10.1101/sqb.2006.71.060. Cold Spring Harb Symp Quant Biol. 2006. PMID: 17381285 Review. - ABC transporters and RNAi in Caenorhabditis elegans.
Timmons LD. Timmons LD. J Bioenerg Biomembr. 2007 Dec;39(5-6):459-63. doi: 10.1007/s10863-007-9121-y. J Bioenerg Biomembr. 2007. PMID: 17994271 Review.
Cited by
- Lack of pairing during meiosis triggers multigenerational transgene silencing in Caenorhabditis elegans.
Leopold LE, Heestand BN, Seong S, Shtessel L, Ahmed S. Leopold LE, et al. Proc Natl Acad Sci U S A. 2015 May 19;112(20):E2667-76. doi: 10.1073/pnas.1501979112. Epub 2015 May 4. Proc Natl Acad Sci U S A. 2015. PMID: 25941370 Free PMC article. - Characterization of virus-encoded RNA interference suppressors in Caenorhabditis elegans.
Guo X, Lu R. Guo X, et al. J Virol. 2013 May;87(10):5414-23. doi: 10.1128/JVI.00148-13. Epub 2013 Mar 6. J Virol. 2013. PMID: 23468484 Free PMC article. - Transgene-Assisted Genetic Screen Identifies rsd-6 and Novel Genes as Key Components of Antiviral RNA Interference in Caenorhabditis elegans.
Long T, Meng F, Lu R. Long T, et al. J Virol. 2018 Aug 16;92(17):e00416-18. doi: 10.1128/JVI.00416-18. Print 2018 Sep 1. J Virol. 2018. PMID: 29950414 Free PMC article. - Germ granules and gene regulation in the Caenorhabditis elegans germline.
Phillips CM, Updike DL. Phillips CM, et al. Genetics. 2022 Mar 3;220(3):iyab195. doi: 10.1093/genetics/iyab195. Genetics. 2022. PMID: 35239965 Free PMC article. Review. - New insights into siRNA amplification and RNAi.
Zhang C, Ruvkun G. Zhang C, et al. RNA Biol. 2012 Aug;9(8):1045-9. doi: 10.4161/rna.21246. Epub 2012 Aug 1. RNA Biol. 2012. PMID: 22858672 Free PMC article.
References
- Amikura, R., K. Hanyu, M. Kashikawa and S. Kobayashi, 2001. Tudor protein is essential for the localization of mitochondrial RNAs in polar granules of Drosophila embryos. Mech. Dev. 107 97–104. - PubMed
- Bardsley, A., K. McDonald and R. E. Boswell, 1993. Distribution of tudor protein in the Drosophila embryo suggests separation of functions based on site of localization. Development 119 207–219. - PubMed
- Brodersen, P., and O. Voinnet, 2006. The diversity of RNA silencing pathways in plants. Trends Genet. 22 268–280. - PubMed
- Brooks, D. R., P. J. Appleford, L. Murray and R. E. Isaac, 2003. An essential role in molting and morphogenesis of Caenorhabditis elegans for ACN-1, a novel member of the angiotensin-converting enzyme family that lacks a metallopeptidase active site. J. Biol. Chem. 278 52340–52346. - PubMed
- Can, A., O. Semiz and O. Cinar, 2005. Bisphenol-A induces cell cycle delay and alters centrosome and spindle microtubular organization in oocytes during meiosis. Mol. Hum. Reprod. 11 389–396. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials