NF-Y dependent epigenetic modifications discriminate between proliferating and postmitotic tissue - PubMed (original) (raw)
NF-Y dependent epigenetic modifications discriminate between proliferating and postmitotic tissue
Aymone Gurtner et al. PLoS One. 2008.
Abstract
The regulation of gene transcription requires posttranslational modifications of histones that, in concert with chromatin remodeling factors, shape the structure of chromatin. It is currently under intense investigation how this structure is modulated, in particular in the context of proliferation and differentiation. Compelling evidence suggests that the transcription factor NF-Y acts as a master regulator of cell cycle progression, activating the transcription of many cell cycle regulatory genes. However, the underlying molecular mechanisms are not yet completely understood. Here we show that NF-Y exerts its effect on transcription through the modulation of the histone "code". NF-Y colocalizes with nascent RNA, while RNA polymerase II is I phosphorylated on serine 2 of the YSPTSPS repeats within its carboxyterminal domain and histones are carrying modifications that represent activation signals of gene expression (H3K9ac and PAN-H4ac). Comparing postmitotic muscle tissue from normal mice and proliferating muscles from mdx mice, we demonstrate by chromatin immunoprecipitation (ChIP) that NF-Y DNA binding activity correlates with the accumulation of acetylated histones H3 and H4 on promoters of key cell cycle regulatory genes, and with their active transcription. Accordingly, p300 is recruited onto the chromatin of NF-Y target genes in a NF-Y-dependent manner, as demonstrated by Re-ChIP. Conversely, the loss of NF-Y binding correlates with a decrease of acetylated histones, the recruitment of HDAC1, and a repressed heterochromatic state with enrichment of histones carrying modifications known to mediate silencing of gene expression (H3K9me3, H3K27me2 and H4K20me3). As a consequence, NF-Y target genes are downregulated in this context. In conclusion, our data indicate a role of NF-Y in modulating the structure and transcriptional competence of chromatin in vivo and support a model in which NF-Y-dependent histone "code" changes contribute to the proper discrimination between proliferating and postmitotic cells in vivo and in vitro.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
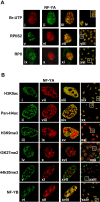
Figure 1. (A) NF-Y is associated with active transcription sites in living cells. After in vivo incorporation of BrUTP (run-on), cells were fixed and endogenous NF-YA (ii) and nascent RNA transcripts (i) were detected by indirect immunofluorescence combined with Confocal Scanning Laser Microscopy by using anti-NF-YA and anti-BrU antibodies.
In the overlay (iii), yellow indicates colocalizations between NF-YA (green) and transcription sites (red). In panels vi and x cells were immunostained with anti-NF-YA, in panels v and ix with anti-RPII CTD repeat YSPTSPS (phospho S2) and anti-total RPII respectively. The majority of NF-YA (red) colocalizes with the activated form of RPII (green)(vii). (B) Cells were immunostained with anti-NF-YA (vii-xii), -acetylated H3K9 (i), -acetylated H4 (ii), -tri-methylated H3K9 (iii), -di-methylated H3K27 (iv), -tri-methylated H4K20 (v) and -NF-YB (vi) antibodies. The majority of NF-YA colocalizes with acetylated (xiii, xiv), but poor colocalization occurs with methylated histones (xv, xvi,xvii). Panel xviii shows the overlay of two subunits of NF-Y, NF-YA (xii) and NF-YB (vi). Panels from xix to xxiv represent a typical optical field of the merge. In figure 1A and 1B confocal analysis of single optical section is shown. The images have been collected with a 60x objective.
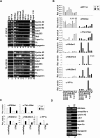
Figure 2. Histone acetylation correlates with NF-Y recruitment onto its target promoters.
(A) ChIPs performed on proliferating (P) and terminally differentiated (TD) C2C12 cells using the indicated antibodies. No antibody was used as control (No Ab). (B) Q-ChIP analysis on proliferating (P) and terminally differentiated (TD) primary myoblasts, performed with the indicated antibodies. The promoters analyzed were: 1) CyclinA2, 2) CyclinB1, 3) CyclinB2, 4) Cdk1, 5) Cdc25C, 6) Dhfr, 7) TopoisomeraseIIα, 8) Tk. (C) Q-ChIP analysis of the Myogenin and MLC promoters performed with the indicated antibodies on proliferating (P) and terminally differentiated (TD) primary myoblasts. On B and C the enrichment of immunoprecipitated promoter fragments relative to the input was done. Rabbit IgG was used as control. (D) RT-PCR amplification of indicated genes was performed on proliferating (P) and terminal differentiated (TD) C2C12.
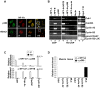
Figure 3. NF-Y recruits p300 onto its target promoters.
(A) Cells were immunosteined with anti-NF-YA (ii, vii), anti-p300 (i), anti-HDAC1 (v) antibodies. Panels iii and vii show the overlay of two proteins, panels iv and viii represents a typical optical field of merge. Confocal analysis of single optical section is shown. (B) ChIPs and re-ChIPs experiments performed with anti-NF-YA, -NF-YB and –p300 antibodies, on proliferating C2C12 cells and stable transfected C2C12 with pmut-CCAATcyclinB2-LUC vector. PCR analysis were performed on the immunoprecipitated DNA samples using specific primers for the indicated promoters and specific primers for the exogenous cyclin B2 mutated promoter. No antibody was used as control (No Ab). (C) Q-Re-ChIP analysis performed on proliferating (P) and terminally differentiated (TD) C2C12 cells using the indicated antibodies on the sames promoters of panel B. (D) Q-ChIP experiments performed with anti-NF-YA, –p300 antibodies, using chromatin prepared from adductor muscles of wt (white bars) and mdx (black bars) of C57BL/10 mice. Specific primers for Cdk1, Cyclin A2, Cyclin B2 and MLC promoters. The values represent the mean of five indipendent experiments performed on triplicate.
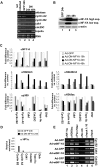
Figure 4. Lack of NF-Y DNA binding activity induces hypermethylation of histones and target gene inactivation.
(A) RT-PCR amplification of the indicated genes from uninfected MEFs (lane 1), MEFs 24hours after infection with Ad-GFP (lane 2), MEFs 24hours and 48hours after infection with Ad-DN-NFYA (lanes 3 and 4). (B) Western blot analysis performed on total lysates from: Mef after 24hours post infection with Ad-GFP (lane1), Mef after 24 hours post infection with Ad-DN-NFYA (lane 2), Mef after 48 hours post infection with Ad-DN-NFYA (lane 3). The extracts were probed with rabbit polyclonal with anti-NFY-A. To normalised protein's loading, the filter was stained with a monoclonal antibody anti-actin protein. (C) Q-ChIP analysis, of the indicated NF-Y target promoters on MEF after 16, 24, 48 hours (h) from Ad-GFP (white bars), or Ad-DN-NFYA after 16 h from infection (dark grey bars), 24 h (black bars), 48 h (grey bars), performed with anti-NFY-A, -p300, -H4K20me3, -H3K9me3 and -H3K9ac antibodies. The fold difference value in each case compares the sample performed after Ad-DN-NFYA infection to the corresponding control sample performed after Ad-GFP infection at the same time point (defined as 1). One of two independent experiments performed in triplicate is represented. (D) Q-ChIP analysis, performed with anti-HP1α antibody, of the indicated NF-Y target promoters on MEF after 24 h after infection with Ad-GFP (white bars) and Ad-DN-NFYA (black bars). It is shown one of two independent experiments performed in triplicate. (E) ChIPs were performed on proliferating MEFs 24 hours after Ad-GFP and wtNF-YA infections. No antibody was used as control (No Ab). PCR analysis were performed on the immunoprecipitated DNA samples with anti-NFY-A, -H3K9ac and -PAN-H4ac-Pan antibodies, using specific primers for the indicate promoters.
Figure 5. Recruitment of euchromatin marks correlates with NF-Y binding in adult tissues.
(A) RT-PCR amplifications of indicate genes were performed on wt and mdx murine muscle total RNAs. Aldolase and Armadillo-X-linked gene were used to normalize. (B) ChIPs performed on wt SDF1 murine muscle tissue using the indicated antibodies. PCR were performed using specific primers for the indicated promoters. (C) Immunohystochemical analysis of wt and mdx muscle tissue using an antibody specific for NF-YA. (D) Q-ChIP analisys of the indicate NF-Y target promoters, performed on muscles of wt (white bars) and mdx (black bars) of C57BL/10 mice, using the indicated antibodies. (E) Q-ChIP analysis of Myosin Light Chain (MLC) muscle specific gene, performed on muscles of wt (white bars) and mdx (black bars) of C57BL/10 mice. The results shown in D and E are the mean of five independent experiments performed in triplicate.
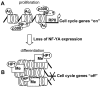
Figure 6. Model proposing the molecular mechanism underlying the transcriptional control of cell cycle-related genes by NF-Y.
Similar articles
- An NF-Y-dependent switch of positive and negative histone methyl marks on CCAAT promoters.
Donati G, Gatta R, Dolfini D, Fossati A, Ceribelli M, Mantovani R. Donati G, et al. PLoS One. 2008 Apr 30;3(4):e2066. doi: 10.1371/journal.pone.0002066. PLoS One. 2008. PMID: 18446193 Free PMC article. - NF-Y is essential for the recruitment of RNA polymerase II and inducible transcription of several CCAAT box-containing genes.
Kabe Y, Yamada J, Uga H, Yamaguchi Y, Wada T, Handa H. Kabe Y, et al. Mol Cell Biol. 2005 Jan;25(1):512-22. doi: 10.1128/MCB.25.1.512-522.2005. Mol Cell Biol. 2005. PMID: 15601870 Free PMC article. - Direct p53 transcriptional repression: in vivo analysis of CCAAT-containing G2/M promoters.
Imbriano C, Gurtner A, Cocchiarella F, Di Agostino S, Basile V, Gostissa M, Dobbelstein M, Del Sal G, Piaggio G, Mantovani R. Imbriano C, et al. Mol Cell Biol. 2005 May;25(9):3737-51. doi: 10.1128/MCB.25.9.3737-3751.2005. Mol Cell Biol. 2005. PMID: 15831478 Free PMC article. - NF-Y and the transcriptional activation of CCAAT promoters.
Dolfini D, Gatta R, Mantovani R. Dolfini D, et al. Crit Rev Biochem Mol Biol. 2012 Jan-Feb;47(1):29-49. doi: 10.3109/10409238.2011.628970. Epub 2011 Nov 3. Crit Rev Biochem Mol Biol. 2012. PMID: 22050321 Review. - Epigenetic landscape of amphetamine and methamphetamine addiction in rodents.
Godino A, Jayanthi S, Cadet JL. Godino A, et al. Epigenetics. 2015;10(7):574-80. doi: 10.1080/15592294.2015.1055441. Epigenetics. 2015. PMID: 26023847 Free PMC article. Review.
Cited by
- Regulating the regulator: a survey of mechanisms from transcription to translation controlling expression of mammalian cell cycle kinase Aurora A.
Cacioppo R, Lindon C. Cacioppo R, et al. Open Biol. 2022 Sep;12(9):220134. doi: 10.1098/rsob.220134. Epub 2022 Sep 7. Open Biol. 2022. PMID: 36067794 Free PMC article. Review. - Specific inhibition of NF-Y subunits triggers different cell proliferation defects.
Benatti P, Dolfini D, Viganò A, Ravo M, Weisz A, Imbriano C. Benatti P, et al. Nucleic Acids Res. 2011 Jul;39(13):5356-68. doi: 10.1093/nar/gkr128. Epub 2011 Mar 16. Nucleic Acids Res. 2011. PMID: 21415014 Free PMC article. - Myo-differentiation reporter screen reveals NF-Y as an activator of PAX3-FOXO1 in rhabdomyosarcoma.
Sroka MW, Skopelitis D, Vermunt MW, Preall JB, El Demerdash O, de Almeida LMN, Chang K, Utama R, Gryder B, Caligiuri G, Ren D, Nalbant B, Milazzo JP, Tuveson DA, Dobin A, Hiebert SW, Stengel KR, Mantovani R, Khan J, Kohli RM, Shi J, Blobel GA, Vakoc CR. Sroka MW, et al. Proc Natl Acad Sci U S A. 2023 Sep 5;120(36):e2303859120. doi: 10.1073/pnas.2303859120. Epub 2023 Aug 28. Proc Natl Acad Sci U S A. 2023. PMID: 37639593 Free PMC article. - MITO-Luc/GFP zebrafish model to assess spatial and temporal evolution of cell proliferation in vivo.
de Latouliere L, Manni I, Ferrari L, Pisati F, Totaro MG, Gurtner A, Marra E, Pacello L, Pozzoli O, Aurisicchio L, Capogrossi MC, Deflorian G, Piaggio G. de Latouliere L, et al. Sci Rep. 2021 Jan 12;11(1):671. doi: 10.1038/s41598-020-79530-5. Sci Rep. 2021. PMID: 33436662 Free PMC article. - The transcription factor NF-Y participates to stem cell fate decision and regeneration in adult skeletal muscle.
Rigillo G, Basile V, Belluti S, Ronzio M, Sauta E, Ciarrocchi A, Latella L, Saclier M, Molinari S, Vallarola A, Messina G, Mantovani R, Dolfini D, Imbriano C. Rigillo G, et al. Nat Commun. 2021 Oct 14;12(1):6013. doi: 10.1038/s41467-021-26293-w. Nat Commun. 2021. PMID: 34650038 Free PMC article.
References
- Ehrenhofer-Murray AE. Chromatin dynamics at DNA replication, transcription and repair. Eur J Biochem. 2004;271(12):2335–2349. - PubMed
- Fischle W, Wang Y, Allis CD. Histone and chromatin cross-talk. Curr Opin Cell Biol. 2003;15(2):172–183. - PubMed
- Li B, Carey M, Workman JL. The Role of Chromatin during Transcription. Cell. 2007;128(4):707–719. - PubMed
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. - PubMed
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129(4):823–37. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous