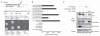Draper-dependent glial phagocytic activity is mediated by Src and Syk family kinase signalling - PubMed (original) (raw)
. 2008 Jun 12;453(7197):935-9.
doi: 10.1038/nature06901. Epub 2008 Apr 23.
Affiliations
- PMID: 18432193
- PMCID: PMC2493287
- DOI: 10.1038/nature06901
Draper-dependent glial phagocytic activity is mediated by Src and Syk family kinase signalling
Jennifer S Ziegenfuss et al. Nature. 2008.
Abstract
The cellular machinery promoting phagocytosis of corpses of apoptotic cells is well conserved from worms to mammals. An important component is the Caenorhabditis elegans engulfment receptor CED-1 (ref. 1) and its Drosophila orthologue, Draper. The CED-1/Draper signalling pathway is also essential for the phagocytosis of other types of 'modified self' including necrotic cells, developmentally pruned axons and dendrites, and axons undergoing Wallerian degeneration. Here we show that Drosophila Shark, a non-receptor tyrosine kinase similar to mammalian Syk and Zap-70, binds Draper through an immunoreceptor tyrosine-based activation motif (ITAM) in the Draper intracellular domain. We show that Shark activity is essential for Draper-mediated signalling events in vivo, including the recruitment of glial membranes to severed axons and the phagocytosis of axonal debris and neuronal cell corpses by glia. We also show that the Src family kinase (SFK) Src42A can markedly increase Draper phosphorylation and is essential for glial phagocytic activity. We propose that ligand-dependent Draper receptor activation initiates the Src42A-dependent tyrosine phosphorylation of Draper, the association of Shark and the activation of the Draper pathway. These Draper-Src42A-Shark interactions are strikingly similar to mammalian immunoreceptor-SFK-Syk signalling events in mammalian myeloid and lymphoid cells. Thus, Draper seems to be an ancient immunoreceptor with an extracellular domain tuned to modified self, and an intracellular domain promoting phagocytosis through an ITAM-domain-SFK-Syk-mediated signalling cascade.
Figures
Figure 1. Shark binds an ITAM in the Draper intracellular domain
a, Draper contains an ITAM domain from Y934-L952 (YXXI-X11-YXXL). The requirements for the five tyrosine residues within and adjacent to this domain (shown) and Src were assayed in the yeast two-hybrid system. b, Lysates from yeast cultures in a were tested in quantitative β-galactosidase (β-Gal) assays. Error bars represent s.e.m.; n = 3; *, P < 0.05. c, S2 cells were transfected with pMT-Myc::Shark and pMT-Drpr-I constructs. Draper immunoprecipitates (IP) were analysed by western blotting (WB) with anti-phosphotyrosine (pTyr), anti-Myc and anti-HA antibodies. Vec., vector.
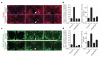
Figure 2. Shark is required for recruitment of Draper and glial membranes to severed axons
a, Control animals (yw;+/UAS-sharkRNAi) and those with glia-specific knockdown of shark (yw;repo-Gal4/UAS-sharkRNAi) were assayed for expression of Draper (red). Glial nuclei were stained with Repo (blue). Left, uninjured; centre, maxillary palp ablation (day 1); right, antennal ablation (day 1). Outlined, example of a maxillary palp-innervated glomerulus; arrow, nerve containing severed maxillary palp ORN axons; open arrowhead, antennal lobe glial cell; boxes, areas quantified in b. b, Quantification of data from a. Error bars represent s.e.m.; n ≥ 10. c, Glial membranes were labelled in control (yw;UAS-GFPS65T/+;repo-Gal4/+) or glial sharkRNAi animals (yw;UAS-GFPS65T/+;repo-Gal4/UAS-sharkRNAi) and assayed for morphology before or after injury (panel order as in a). Outlined, maxillary palp-innervated glomerulus; arrow, nerve containing severed maxillary palp ORN axons; boxes, areas used to quantify glial hypertrophy in d. d, Quantification data from c. Error bars represent s.e.m.; n ≥ 10.
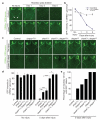
Figure 3. Shark is required for glial clearance of severed axons from the CNS
a, The axons of OR85e-expressing ORNs were labelled with mCD8::GFP in control (yw;OR85e-mCD8::GFP/+;repo-Gal4/+) and glial sharkRNAi (yw;OR85e-mCD8::GFP/+;repo-Gal4/UAS-sharkRNAi) animals, maxillary palps were ablated, and the clearance of severed ORN axons from the CNS was assayed with anti-GFP antibody stains (green). Maxillary nerves are indicated (arrowheads). b, Quantification of data from a. Error bars represent s.e.m.; n ≥ 10. c, OR85e-expressing ORN axons were labelled in control (yw;OR85e-mCD8::GFP/+) animals and in shark1 or draperΔ5 null mutant backgrounds, maxillary palps were ablated, and clearance was assayed at 5 days. d, Quantification of data from c. Error bars represent s.e.m.; n ≥ 10; *, P < 0.05; **, P < 0.001; ***, P < 0.0001. e, Quantification of brain hemispheres containing GFP-labelled ORN axonal debris along the maxillary nerve for genotypes described in c.
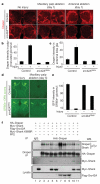
Figure 4. Src42Aa is required for glial responses to axon injury and modulates Draper phosphorylation status
a, Control animals (yw;UAS-src42ARNAi/+, no driver) and those with glia-specific knockdown of src42A (yw;UAS-src42ARNAi/+;repo-Gal4/+) were assayed for injury-induced changes in glial Draper expression and for recruitment of Draper to severed axons (red). b, c, Quantification of data from a for palp-innervated glomeruli (b) and antennal lobe glia (c). Error bars represent s.e.m.; n ≥ 10. d, The axons of OR85e-expressing ORNs were labelled with mCD8::GFP in control (yw;OR85e-mCD8::GFP/+) and glial src42ARNAi (yw;OR85e-mCD8::GFP/UAS-src42ARNAi;repo-Gal4/+) animals, maxillary palps were ablated, and the clearance of severed ORN axons from the CNS was assayed with anti-GFP antibody stains (green) 5 days after injury. e, Quantification of data from d. Error bars represent s.e.m.; n ≥ 10. f, S2 cells were co-transfected with pMT-HA::Draper, pMT-Myc::Shark, pMT-Flag::Src42A, pMT-Myc::Shark K698R (kinase-dead) and pMT vector. After transfection and expression, some cells were incubated for 60 min with the SFK inhibitor PP2 (10 μM) before cell lysis. Anti-Draper and IgG control immunoprecipitates (IP) from cells were analysed by SDS-PAGE and western blotted (WB) with antibodies against pTyr, Draper, Myc and Flag.
Similar articles
- Jedi-1 and MEGF10 signal engulfment of apoptotic neurons through the tyrosine kinase Syk.
Scheib JL, Sullivan CS, Carter BD. Scheib JL, et al. J Neurosci. 2012 Sep 19;32(38):13022-31. doi: 10.1523/JNEUROSCI.6350-11.2012. J Neurosci. 2012. PMID: 22993420 Free PMC article. - Draper/CED-1 mediates an ancient damage response to control inflammatory blood cell migration in vivo.
Evans IR, Rodrigues FS, Armitage EL, Wood W. Evans IR, et al. Curr Biol. 2015 Jun 15;25(12):1606-12. doi: 10.1016/j.cub.2015.04.037. Epub 2015 May 28. Curr Biol. 2015. PMID: 26028435 Free PMC article. - Axon degeneration induces glial responses through Draper-TRAF4-JNK signalling.
Lu TY, MacDonald JM, Neukomm LJ, Sheehan AE, Bradshaw R, Logan MA, Freeman MR. Lu TY, et al. Nat Commun. 2017 Feb 6;8:14355. doi: 10.1038/ncomms14355. Nat Commun. 2017. PMID: 28165006 Free PMC article. - Protein tyrosine kinase, syk: a key player in phagocytic cells.
Tohyama Y, Yamamura H. Tohyama Y, et al. J Biochem. 2009 Mar;145(3):267-73. doi: 10.1093/jb/mvp001. Epub 2009 Jan 4. J Biochem. 2009. PMID: 19124456 Review. - Protein tyrosine kinase Syk in mast cell signaling.
Siraganian RP, Zhang J, Suzuki K, Sada K. Siraganian RP, et al. Mol Immunol. 2002 Sep;38(16-18):1229-33. doi: 10.1016/s0161-5890(02)00068-8. Mol Immunol. 2002. PMID: 12217388 Review.
Cited by
- Eaten alive: novel insights into autophagy from multicellular model systems.
Zhang H, Baehrecke EH. Zhang H, et al. Trends Cell Biol. 2015 Jul;25(7):376-87. doi: 10.1016/j.tcb.2015.03.001. Epub 2015 Apr 7. Trends Cell Biol. 2015. PMID: 25862458 Free PMC article. Review. - Microglial immunoreceptor tyrosine-based activation and inhibition motif signaling in neuroinflammation.
Linnartz B, Wang Y, Neumann H. Linnartz B, et al. Int J Alzheimers Dis. 2010 Jun 22;2010:587463. doi: 10.4061/2010/587463. Int J Alzheimers Dis. 2010. PMID: 20721346 Free PMC article. - trim-21 promotes proteasomal degradation of CED-1 for apoptotic cell clearance in C. elegans.
Yuan L, Li P, Jing H, Zheng Q, Xiao H. Yuan L, et al. Elife. 2022 Aug 5;11:e76436. doi: 10.7554/eLife.76436. Elife. 2022. PMID: 35929733 Free PMC article. - Syk-MyD88 Axis Is a Critical Determinant of Inflammatory-Response in Activated Macrophages.
Yi YS, Kim HG, Kim JH, Yang WS, Kim E, Jeong D, Park JG, Aziz N, Kim S, Parameswaran N, Cho JY. Yi YS, et al. Front Immunol. 2021 Dec 23;12:767366. doi: 10.3389/fimmu.2021.767366. eCollection 2021. Front Immunol. 2021. PMID: 35003083 Free PMC article. - Sialic acid on the neuronal glycocalyx prevents complement C1 binding and complement receptor-3-mediated removal by microglia.
Linnartz B, Kopatz J, Tenner AJ, Neumann H. Linnartz B, et al. J Neurosci. 2012 Jan 18;32(3):946-52. doi: 10.1523/JNEUROSCI.3830-11.2012. J Neurosci. 2012. PMID: 22262892 Free PMC article.
References
- Keegan K, Cooper JA. Use of the two hybrid system to detect the association of the protein-tyrosine-phosphatase, SHPTP2, with another SH2-containing protein, Grb7. Oncogene. 1996;12:1537–1544. - PubMed
- Lioubin MN, et al. p150Ship, a signal transduction molecule with inositol polyphosphate-5-phosphatase activity. Genes Dev. 1996;10:1084–1095. - PubMed
- Li W, Yeung YG, Stanley ER. Tyrosine phosphorylation of a common 57-kDa protein in growth factor-stimulated and-transformed cells. J. Biol. Chem. 1991;266:6808–6814. - PubMed
- Zhou Z, Hartwieg E, Horvitz HR. CED-1 is a transmembrane receptor that mediates cell corpse engulfment in C. elegans. Cell. 2001;104:43–56. - PubMed
- Freeman MR, Delrow J, Kim J, Johnson E, Doe CQ. Unwrapping glial biology. Gcm target genes regulating glial development, diversification, and function. Neuron. 2003;38:567–580. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 1R01NS053538/NS/NINDS NIH HHS/United States
- R37 CA026504-30/CA/NCI NIH HHS/United States
- 1R01CA26504/CA/NCI NIH HHS/United States
- R01 CA026504/CA/NCI NIH HHS/United States
- R37 CA026504/CA/NCI NIH HHS/United States
- 1R01GM55293/GM/NIGMS NIH HHS/United States
- R01 NS053538/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous