Conditional loss of Dicer disrupts cellular and tissue morphogenesis in the cortex and hippocampus - PubMed (original) (raw)
Comparative Study
Conditional loss of Dicer disrupts cellular and tissue morphogenesis in the cortex and hippocampus
Tigwa H Davis et al. J Neurosci. 2008.
Abstract
To investigate the role of Dicer and microRNAs in the mammalian CNS, we used mice in which the second RNase III domain of Dicer was conditionally floxed. Conditional Dicer mice were bred with mice expressing an alpha-calmodulin kinase II Cre to selectively inactivate Dicer in excitatory forebrain neurons in vivo. Inactivation of Dicer results in an array of phenotypes including microcephaly, reduced dendritic branch elaboration, and large increases in dendritic spine length with no concomitant change in spine density. Microcephaly is likely caused by a 5.5-fold increase in early postnatal apoptosis in these animals as determined by active caspase-3 and TUNEL (terminal deoxynucleotidyl transferase-mediated biotinylated UTP nick end labeling) staining in the cortex. Loss of Dicer function had no measurable effect on cortical lamination as determined by in situ hybridization, suggesting that microcephaly is not caused by defects in neuronal migration. Together, these results illustrate the in vivo significance of Dicer and miRNAs in the mammalian CNS and provide additional support for previous in vitro studies indicating that misregulation of this pathway may result in gross abnormalities in cell number and function that may contribute to a variety of neurological disorders.
Figures
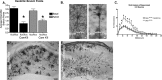
Figure 1.
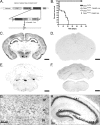
Cre recombination occurs throughout the forebrain and results in early death. A, Dicer targeting construct. The second RNase III domain of Dicer was flanked with Lox P sites. After Cre recombination at embryonic day 15.5, Dicer is rendered inactive, leading to a failure to produce microRNAs in excitatory forebrain postmitotic neurons. B, Kaplan–Meier plot of survival demonstrates that 40% of Dicerflox/flox; CaMKII Cre (n = 26; open triangle) animals die by postnatal day 2 and that the remaining animals die by the end of postnatal day 21, whereas other genotypes persist into adulthood (Dicerflox/flox, n = 20, open square; Dicerflox/wt, n = 20, open circle; Dicerflox/wt; CaMKII Cre, n = 20, closed square). C–H, Floxed Dicer mice were crossed to α-CaMKII Cre- and RosaR26R-expressing mice to visualize brain regions in which Dicer was removed. C, Cre-recombined cells at postnatal day 21 in a Dicerflox/wt; CaMKII Cre; R26R +/− mouse. Recombination is high throughout all cortical layers as well as throughout the pyramidal cell layer of the hippocampus. Little expression/recombination is observed in the thalamus. D, Cre recombination at postnatal day 21 in a Dicerflox/wt; CaMKII Cre; R26R +/+ mouse demonstrating the specificity of X-gal staining. E, X-gal staining at the level of the cerebellum in a Dicerflox/wt; CaMKII Cre; R26R +/− mouse illustrating forebrain specificity of the CaMKII Cre line. F, Similar to the image in D, there is a lack of X-gal staining in the cerebellum in Dicerflox/wt; CaMKII Cre; R26R +/+ mice. G, Higher magnification of cortical layers IV–VI. Recombination occurs in most but not all cortical neurons. H, Higher magnification of the hippocampal formation in a Dicerflox/wt; CaMKII Cre mouse illustrating that recombination occurs throughout the dentate gyrus and CA1–3 regions, but not in strata, where interneurons and glia predominate. Scale bars: C–F, 1 mm. RBD, RNA binding domain; FRT, flipase recognition target.
Figure 2.
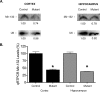
MicroRNA levels are decreased in Dicer mutant mice. A, Northern blot analysis for mir-132 at P15 demonstrates a decrease in miRNA levels in the cortex and hippocampus of Dicerflox/flox; CaMKII Cre mice. B, qRT-PCR analysis of mir-132 levels in the cortex and hippocampus of Dicer mutant mice at P21 demonstrates that miRNA levels continue to decrease over time. U6 was used as a loading control in all experiments. Five micrograms of total RNA were loaded per well for cortical and hippocampal mir-132 Northern blot analysis. Twenty nanograms of total RNA were used to generate cDNA for qRT-PCR analysis. Error bars represent the SEM. Values are statistically significant using a one-way ANOVA (*p < 0.001) with a Tukey's multiple comparison-post test.
Figure 3.
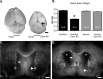
Dicer mutant animals show decreases in brain weight and abnormally large ventricles. A, Brains of Dicer mutant animals are consistently smaller than their wild-type littermates. The arrow highlights increased exposure of superior colliculus in Dicer mutant animals. B, Quantification of brain weight (N indicates number of animals). C, D, Dicerflox/flox (C) and Dicerflox/flox; CaMKII Cre +/− (D) mice immunostained for neuropilin-2 demonstrate substantial differences in ventricular size as well as axonal pathfinding abnormalities (arrows in C and D). Arrowheads illustrate differences in anterior commissure. Scale bars: A, 1 mm; C, D, 500 μm. In B, error bars represent SEM. Values are statistically significant (*p < 0.001) using a one-way ANOVA with a Tukey's multiple comparison post test.
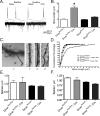
Figure 4.
Dicer inactivation leads to increased cortical apoptosis in vivo. A, Quantification of active caspase-3 staining in Dicerflox/wt; CaMKII +/− and Dicerflox/flox; CaMKII +/− at postnatal day 0 (Dicerflox/flox; CaMKII Cre +/−, n = 25, N = 3; Dicerflox/wt; CaMKII Cre +/−, n = 24, N = 3). For negative controls, n = 12 and N = 3. N indicates the number of animals, and n the number of sections analyzed. B, Immunostaining for the active (cleaved) form of caspase-3 illustrating neurons adjacent to the lateral ventricle undergoing programmed cell death. Inset, Apoptotic layer 5 cortical neuron (same section) showing blebbing characteristic of programmed cell death. C, TUNEL staining of Dicerflox/flox (top) and Dicerflox/flox; CaMKII Cre animals (bottom) at postnatal day 0 indicating that Dicer mutant animals have multifold increases in cortical apoptosis with the majority of apoptosis occurring at or around the lateral ventricles in the cortex of Dicer mutant animals. Positive controls were first treated with 1 μg/μl DNase I in 1× TBS-Tween 20/MgSO4 for 20 min at room temperature to cause DNA damage and then processed with other sections. In A and C, negative controls refer to samples untreated with an anti-caspase-3 antibody or terminal deoxynucleotidyl transferase, respectively. Scale bars: B, 10 μm; C, 100 μm. In A, error bars represent SEM. Values are statistically significant using a one-way ANOVA (*p < 0.001) with a Tukey's multiple comparison post test.
Figure 5.
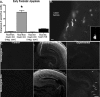
Dendritic branching in hippocampal neurons is impaired in Dicer mutant animals. A, Quantification of basal and apical dendritic branch points in Dicerflox/flox and Dicerflox/flox; CaMKII +/− Cre animals demonstrate decreases in the number of dendritic branch points. B, Example of Golgi-stained hippocampal CA1 neurons from Dicerflox/flox and Dicerflox/flox; CaMKII +/− animals illustrating differences in basal and apical branching. C, Sholl analysis of Dicerflox/flox and Dicerflox/flox; CaMKII +/− animals illustrates substantial differences in dendritic complexity (Dicerflox/flox, n = 24, N = 3; Dicerflox/flox; CaMKII +/−, n = 24, N = 3). D, Golgi-stained Dicerflox/flox hippocampus with stereotyped architecture including a well defined dentate gyrus and CA1–3 region. E, Golgi-stained Dicerflox/flox; CaMKII +/− Cre hippocampus outlined in white illustrates decreased size relative to that shown in E (images taken at same magnification). Abnormal hippocampal architecture observed in Dicerflox/flox; CaMKII +/− Cre mice makes distinct layers of the CA region difficult to discern. The white arrow highlights loss of the dentate gyrus. Images were taken at 4× in D and E. n is the number of neurons, N the number of animals. Scale bars: B, 10 μm; E, F, 100 μm. Error bars represent SEM. Values in A are statistically significant using a one-way ANOVA (*p < 0.001) with a Tukey's multiple comparison post test. Values in C are significant using an unpaired two-tailed t test (*p < 0.05).
Figure 6.
Inactivation of Dicer in hippocampal CA1 neurons leads to increased apical dendritic spine length but not density in vivo. A, Spontaneous action potentials and synaptic activity from Dicer mutant animals at postnatal day 15 indicate that dendritic spines are functional. B, Mean dendritic spine length for Dicerflox/flox, 0.98 ± 0.04 (n = 245, N = 2); Dicerflox/flox; CaMKII +/−, 2.40 ± 0.23 (n = 117, N = 2); and Dicerflox/wt, 0.94 ± 0.03 (n = 236, N = 3); Dicerflox/wt; CaMKII +/−, 1.26 ± 0.08 (n = 141, N = 3). C, Example of dendritic spines in Dicerflox/flox; CaMKII +/− Cre −/−, and Dicerflox/wt; CaMKII +/− Cre +/− animals. Dendritic spines of Dicerflox/flox; CaMKII +/− Cre (as shown in C, left) were observed up to 12 μm in length. D, Cumulative distribution of apical dendritic spine length in each Dicer genotype. Loss of microRNAs results in one-third of dendritic spines being greater in length than the longest measured dendritic spine in other Dicer genotypes. E, F, Basal and apical dendritic spine density in CA1 hippocampal neurons at postnatal day 21 in Golgi-stained neurons. The number of spines per 50 μm dendrite were counted by three blind observers, and the numbers were averaged. Criterion for inclusion included dendrites of 75 μm or longer. For analysis, spines were counted on basal or apical dendrites starting 25 μm from the cell body for a length of 50 μm. Analysis performed on two to three dendrites per cell in each condition with four to five cells measured. Three to four animals were used per condition. N is the number of animals, and n the number of cells. Scale bars, 2 μm. Error bars represent SEM. Values in B are statistically significant (*p < 0.001) using a one-way ANOVA with a Tukey's post test.
Similar articles
- Age-dependent neuron loss is associated with impaired adult neurogenesis in forebrain neuron-specific Dicer conditional knockout mice.
Cheng S, Zhang C, Xu C, Wang L, Zou X, Chen G. Cheng S, et al. Int J Biochem Cell Biol. 2014 Dec;57:186-96. doi: 10.1016/j.biocel.2014.10.029. Epub 2014 Nov 3. Int J Biochem Cell Biol. 2014. PMID: 25448413 - Global microRNA expression is essential for murine mast cell development in vivo.
Oh SY, Brandal S, Kapur R, Zhu Z, Takemoto CM. Oh SY, et al. Exp Hematol. 2014 Oct;42(10):919-23.e1. doi: 10.1016/j.exphem.2014.07.266. Epub 2014 Sep 6. Exp Hematol. 2014. PMID: 25201754 Free PMC article. - Dicer deficiency reveals microRNAs predicted to control gene expression in the developing adrenal cortex.
Krill KT, Gurdziel K, Heaton JH, Simon DP, Hammer GD. Krill KT, et al. Mol Endocrinol. 2013 May;27(5):754-68. doi: 10.1210/me.2012-1331. Epub 2013 Mar 21. Mol Endocrinol. 2013. PMID: 23518926 Free PMC article. - Deletion of astroglial Dicer causes non-cell-autonomous neuronal dysfunction and degeneration.
Tao J, Wu H, Lin Q, Wei W, Lu XH, Cantle JP, Ao Y, Olsen RW, Yang XW, Mody I, Sofroniew MV, Sun YE. Tao J, et al. J Neurosci. 2011 Jun 1;31(22):8306-19. doi: 10.1523/JNEUROSCI.0567-11.2011. J Neurosci. 2011. PMID: 21632951 Free PMC article. - Role of Dicer in female fertility.
Luense LJ, Carletti MZ, Christenson LK. Luense LJ, et al. Trends Endocrinol Metab. 2009 Aug;20(6):265-72. doi: 10.1016/j.tem.2009.05.001. Epub 2009 Jul 29. Trends Endocrinol Metab. 2009. PMID: 19646895 Free PMC article. Review.
Cited by
- Gene network and pathway analysis of mice with conditional ablation of Dicer in post-mitotic neurons.
Dorval V, Smith PY, Delay C, Calvo E, Planel E, Zommer N, Buée L, Hébert SS. Dorval V, et al. PLoS One. 2012;7(8):e44060. doi: 10.1371/journal.pone.0044060. Epub 2012 Aug 27. PLoS One. 2012. PMID: 22952873 Free PMC article. - Beneficial effects of miR-132/212 deficiency in the zQ175 mouse model of Huntington's disease.
Nateghi B, Keraudren R, Boulay G, Bazin M, Goupil C, Canet G, Loiselle A, St-Amour I, Planel E, Soulet D, Hébert SS. Nateghi B, et al. Front Neurosci. 2024 Aug 7;18:1421680. doi: 10.3389/fnins.2024.1421680. eCollection 2024. Front Neurosci. 2024. PMID: 39170678 Free PMC article. - Advances in microRNA experimental approaches to study physiological regulation of gene products implicated in CNS disorders.
Long JM, Lahiri DK. Long JM, et al. Exp Neurol. 2012 Jun;235(2):402-18. doi: 10.1016/j.expneurol.2011.12.043. Epub 2012 Jan 5. Exp Neurol. 2012. PMID: 22245616 Free PMC article. Review. - Reduced mature microRNA levels in association with dicer loss in human temporal lobe epilepsy with hippocampal sclerosis.
McKiernan RC, Jimenez-Mateos EM, Bray I, Engel T, Brennan GP, Sano T, Michalak Z, Moran C, Delanty N, Farrell M, O'Brien D, Meller R, Simon RP, Stallings RL, Henshall DC. McKiernan RC, et al. PLoS One. 2012;7(5):e35921. doi: 10.1371/journal.pone.0035921. Epub 2012 May 15. PLoS One. 2012. PMID: 22615744 Free PMC article. - Non-coding RNA in neural function, disease, and aging.
Szafranski K, Abraham KJ, Mekhail K. Szafranski K, et al. Front Genet. 2015 Mar 9;6:87. doi: 10.3389/fgene.2015.00087. eCollection 2015. Front Genet. 2015. PMID: 25806046 Free PMC article. Review.
References
- Arlotta P, Molyneaux BJ, Chen J, Inoue J, Kominami R, Macklis JD. Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron. 2005;45:207–221. - PubMed
- Bagnard D, Lohrum M, Uziel D, Puschel AW, Bolz J. Semaphorins act as attractive and repulsive guidance signals during the development of cortical projections. Development. 1998;125:5043–5053. - PubMed
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. - PubMed
- Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ. Dicer is essential for mouse development. Nat Genet. 2003;35:215–217. - PubMed
- Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nat Struct Mol Biol. 2006;13:1097–1101. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- T32EY07120/EY/NEI NIH HHS/United States
- R03 DA022201/DA/NIDA NIH HHS/United States
- T32 EY007120/EY/NEI NIH HHS/United States
- R21-MH083090/MH/NIMH NIH HHS/United States
- R03-DA022201/DA/NIDA NIH HHS/United States
- R03 DA022201-01/DA/NIDA NIH HHS/United States
- EY002162/EY/NEI NIH HHS/United States
- AS1792/AS/Autism Speaks/United States
- R03 DA022201-02/DA/NIDA NIH HHS/United States
- R21 MH083090/MH/NIMH NIH HHS/United States
- P30 EY002162/EY/NEI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials