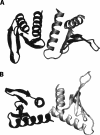RDC-assisted modeling of symmetric protein homo-oligomers - PubMed (original) (raw)
RDC-assisted modeling of symmetric protein homo-oligomers
Xu Wang et al. Protein Sci. 2008 May.
Abstract
Protein oligomerization serves an important function in biological processes, yet solving structures of protein oligomers has always been a challenge. For solution NMR, the challenge arises both from the increased size of these systems and, in the case of homo-oligomers, from ambiguities in assignment of intra- as opposed to intersubunit NOEs. In this study, we present a residual dipolar coupling (RDC)-assisted method for constructing models of homo-oligomers with purely rotational symmetry. Utilizing the fact that one of the principal axes of the tensor describing the alignment needed for RDC measurement is always parallel to the oligomer symmetry axis, it is possible to greatly restrict possible models for the oligomer. Here, it is shown that, if the monomer structure is known, all allowed dimer models can be constructed using a grid search algorithm and evaluated based on RDC simulations and the quality of the interface between the subunits. Using the Bacillus subtilis protein YkuJ as an example, it is shown that the evaluation criteria based on just two sets of NH RDCs are very selective and can unambiguously produce a model in good agreement with an existing X-ray structure of YkuJ.
Figures
Figure 1.
Schematic diagram of the two possible dimer models from the X-ray structure of YkuJ (SR360) (PDB access code 2FFG). (A) The dimer formed in the asymmetric unit. (B) The dimer formed by two molecules across the asymmetric unit. Subunit with the same shade represents the same molecule in unit cell.
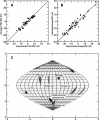
Figure 2.
(A) Correlation between experimental RDCs collected in a PEG alignment medium and the back-calculated RDCs using dimer model B from the X-ray structure. (B) Correlation between experimental RDCs collected in phage alignment medium and the back-calculated RDCs using dimer model B from the X-ray structure. (C) Sauson–Flamsteed projection of the orientation of the PEG and phage alignment tensor principal axes frames in the PDB coordinate. Each horizontal grid represents 20° and each vertical grid represents 10°. (Black dot) Orientation of the crystal structure symmetry axis.
Figure 3.
Algorithm for constructing all possible dimer models given the symmetry axis using a grid search algorithm.
Figure 4.
Surface plots of the combined score of dimer models generated using the _X_-axis of the PEG alignment tensor (A), the phage alignment tensor (B), and the consensus orientation (C) as the symmetry axis. During the search, the center of mass of one subunit is fixed at the point (35 Å, 35 Å). The translation refers to the coordinate of the center of mass of the second subunit. Models with VDW energy higher than the median value or RDC correlation <0.85 or residual pairing score <0 are given a score of 0. The score is indicated by the color of each grid point: (black) 0, (yellow) 1. The color gradient for the score is also shown on the right of each plot.
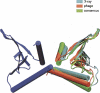
Figure 5.
Superimposition of the best models generated by the phage alignment tensor and the consensus searches with dimer model B from the X-ray structure. Chain A in all models are superimposed and are represented by the blue subunit. The best model for each search is chosen by plotting the grid positions of the top 30 models in each search and choosing the model closest to the center. Phage alignment tensor is from grid position (47 Å, 59 Å). Consensus axis search model is from grid position (37 Å, 63 Å).
Similar articles
- A geometric arrangement algorithm for structure determination of symmetric protein homo-oligomers from NOEs and RDCs.
Martin JW, Yan AK, Bailey-Kellogg C, Zhou P, Donald BR. Martin JW, et al. J Comput Biol. 2011 Nov;18(11):1507-23. doi: 10.1089/cmb.2011.0173. Epub 2011 Oct 28. J Comput Biol. 2011. PMID: 22035328 Free PMC article. - The use of residual dipolar coupling in studying proteins by NMR.
Chen K, Tjandra N. Chen K, et al. Top Curr Chem. 2012;326:47-67. doi: 10.1007/128_2011_215. Top Curr Chem. 2012. PMID: 21952837 Free PMC article. Review. - A graphical method for analyzing distance restraints using residual dipolar couplings for structure determination of symmetric protein homo-oligomers.
Martin JW, Yan AK, Bailey-Kellogg C, Zhou P, Donald BR. Martin JW, et al. Protein Sci. 2011 Jun;20(6):970-85. doi: 10.1002/pro.620. Epub 2011 Apr 27. Protein Sci. 2011. PMID: 21413097 Free PMC article. - Top-down approach in protein RDC data analysis: de novo estimation of the alignment tensor.
Chen K, Tjandra N. Chen K, et al. J Biomol NMR. 2007 Aug;38(4):303-13. doi: 10.1007/s10858-007-9168-4. Epub 2007 Jun 26. J Biomol NMR. 2007. PMID: 17593526 - New opportunities for tensor-free calculations of residual dipolar couplings for the study of protein dynamics.
Montalvao R, Camilloni C, De Simone A, Vendruscolo M. Montalvao R, et al. J Biomol NMR. 2014 Apr;58(4):233-8. doi: 10.1007/s10858-013-9801-3. Epub 2014 Jan 30. J Biomol NMR. 2014. PMID: 24477919 Review.
Cited by
- Post-translational S-nitrosylation is an endogenous factor fine tuning the properties of human S100A1 protein.
Lenarčič Živković M, Zaręba-Kozioł M, Zhukova L, Poznański J, Zhukov I, Wysłouch-Cieszyńska A. Lenarčič Živković M, et al. J Biol Chem. 2012 Nov 23;287(48):40457-70. doi: 10.1074/jbc.M112.418392. Epub 2012 Sep 18. J Biol Chem. 2012. PMID: 22989881 Free PMC article. - A geometric arrangement algorithm for structure determination of symmetric protein homo-oligomers from NOEs and RDCs.
Martin JW, Yan AK, Bailey-Kellogg C, Zhou P, Donald BR. Martin JW, et al. J Comput Biol. 2011 Nov;18(11):1507-23. doi: 10.1089/cmb.2011.0173. Epub 2011 Oct 28. J Comput Biol. 2011. PMID: 22035328 Free PMC article. - The use of residual dipolar coupling in studying proteins by NMR.
Chen K, Tjandra N. Chen K, et al. Top Curr Chem. 2012;326:47-67. doi: 10.1007/128_2011_215. Top Curr Chem. 2012. PMID: 21952837 Free PMC article. Review. - Automated NMR Assignment and Protein Structure Determination using Sparse Dipolar Coupling Constraints.
Donald BR, Martin J. Donald BR, et al. Prog Nucl Magn Reson Spectrosc. 2009 Aug 1;55(2):101-127. doi: 10.1016/j.pnmrs.2008.12.001. Prog Nucl Magn Reson Spectrosc. 2009. PMID: 20160991 Free PMC article. No abstract available. - Solution structure of human growth arrest and DNA damage 45alpha (Gadd45alpha) and its interactions with proliferating cell nuclear antigen (PCNA) and Aurora A kinase.
Sánchez R, Pantoja-Uceda D, Prieto J, Diercks T, Marcaida MJ, Montoya G, Campos-Olivas R, Blanco FJ. Sánchez R, et al. J Biol Chem. 2010 Jul 16;285(29):22196-201. doi: 10.1074/jbc.M109.069344. Epub 2010 May 11. J Biol Chem. 2010. PMID: 20460379 Free PMC article.
References
- Al-Hashimi, H.M., Bolon, P.J., Prestegard, J.H. Molecular symmetry as an aid to geometry determination in ligand protein complexes. J. Magn. Reson. 2000;142:153–158. - PubMed
- Al-Hashimi, H.M., Majumdar, A., Gorin, A., Kettani, A., Skripkin, E., Patel, D.J. Field- and phage-induced dipolar couplings in a homodimeric DNA quadruplex: Relative orientation of G · (C-A) triad and G-tetrad motifs and direct determination of C2 symmetry axis orientation. J. Am. Chem. Soc. 2001;123:633–640. - PubMed
- Ali, M.H., Imperiali, B. Protein oligomerization: How and why. Bioorg. Med. Chem. 2005;13:5013–5020. - PubMed
- Bax, A., Grishaev, A. Weak alignment NMR: A hawk-eyed view of biomolecular structure. Curr. Opin. Struct. Biol. 2005;15:563–570. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources