A new protein linear motif benchmark for multiple sequence alignment software - PubMed (original) (raw)
A new protein linear motif benchmark for multiple sequence alignment software
Emmanuel Perrodou et al. BMC Bioinformatics. 2008.
Abstract
Background: Linear motifs (LMs) are abundant short regulatory sites used for modulating the functions of many eukaryotic proteins. They play important roles in post-translational modification, cell compartment targeting, docking sites for regulatory complex assembly and protein processing and cleavage. Methods for LM detection are now being developed that are strongly dependent on scores for motif conservation in homologous proteins. However, most LMs are found in natively disordered polypeptide segments that evolve rapidly, unhindered by structural constraints on the sequence. These regions of modular proteins are difficult to align using classical multiple sequence alignment programs that are specifically optimised to align the globular domains. As a consequence, poor motif alignment quality is hindering efforts to detect new LMs.
Results: We have developed a new benchmark, as part of the BAliBASE suite, designed to assess the ability of standard multiple alignment methods to detect and align LMs. The reference alignments are organised into different test sets representing real alignment problems and contain examples of experimentally verified functional motifs, extracted from the Eukaryotic Linear Motif (ELM) database. The benchmark has been used to evaluate and compare a number of multiple alignment programs. With distantly related proteins, the worst alignment program correctly aligns 48% of LMs compared to 73% for the best program. However, the performance of all the programs is adversely affected by the introduction of other sequences containing false positive motifs. The ranking of the alignment programs based on LM alignment quality is similar to that observed when considering full-length protein alignments, however little correlation was observed between LM and overall alignment quality for individual alignment test cases.
Conclusion: We have shown that none of the programs currently available is capable of reliably aligning LMs in distantly related sequences and we have highlighted a number of specific problems. The results of the tests suggest possible ways to improve program accuracy for difficult, divergent sequences.
Figures
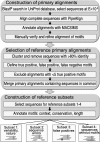
Figure 1
BAliBASE Reference Set 9 construction protocol. Flow-chart showing the 3 major steps of the protocol used to construct the BAliBASE Reference Set 9.
Figure 2
Example alignment in BAliBASE Reference Set 9. Part of an alignment, showing the MOD_PKM_1 ELM regular expression (R.R.. [ST]...), with examples of true positive, false positive and false negative motifs. The last three sequences do not contain any examples of the motif and cannot be aligned unambiguously in the region of the motif instance.
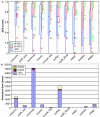
Figure 3
Program SPS scores at different levels of overall sequence similarity. a) Box plots of the SPS scores obtained by the different alignment programs in subset 1, showing the extreme observations (stars or circles), lower quartile, median, upper quartile, and largest observation in each similarity category. b) Execution times in seconds required to construct all the multiple alignments in Subset 1. Programs are displayed in the order of the Friedman test using the SPS scores for group V11 (additional file 1), with the highest scoring program on the left.
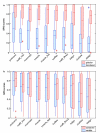
Figure 4
Program SPS scores depending on different motif characteristics. Box plots of the SPS scores obtained by the different alignment programs in subset 1, group V11 (<20% identity) under different conditions. The boxplots indicate the extreme observations (stars), lower quartile, median, upper quartile, and largest observation. Significant differences, according to a Wilcoxon signed ranks test (p < 0.05), are indicated by an asterix on the x-axis. P-values for the Wilcoxon tests are available in additional file 1, table 2. a) SPS scores for motifs found in globular domains versus disordered regions. b) SPS scores for motifs with a conserved residue versus variable motifs.
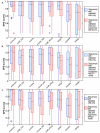
Figure 5
Program SPS scores after inclusion of sequences without validated motifs. Box plots of the SPS scores obtained by the different alignment programs under different conditions, showing the extreme observations (stars or circles), lower quartile, median, upper quartile, and largest observation. Significant differences, according to a Wilcoxon signed ranks test (p < 0.05), are indicated by an asterix on the x-axis. P-values for the Wilcoxon tests are available in additional file 1, table 3. a) SPS scores for alignments of sequences with validated motifs only compared to alignments including sequences with errors. b) SPS scores for alignments of sequences with validated motifs only compared to alignments including sequences containing false positive (FP) motifs. c) SPS scores for alignments of sequences with validated motifs only compared to alignments including sequences that do not contain any examples of the motif.
Figure 6
Program NorMD scores at different levels of overall sequence similarity. Box plots of the NorMD scores, representing the alignment quality of the full length alignment, obtained by the different alignment programs for the different similarity categories in subset 1. The boxplots indicate the extreme observations (stars), lower quartile, median, upper quartile, and largest observation. Programs are displayed in the order of the Friedman test using the NorMD scores for group V11 (additional file 1), with the highest scoring program on the left.
Similar articles
- BAliBASE 3.0: latest developments of the multiple sequence alignment benchmark.
Thompson JD, Koehl P, Ripp R, Poch O. Thompson JD, et al. Proteins. 2005 Oct 1;61(1):127-36. doi: 10.1002/prot.20527. Proteins. 2005. PMID: 16044462 - OXBench: a benchmark for evaluation of protein multiple sequence alignment accuracy.
Raghava GP, Searle SM, Audley PC, Barber JD, Barton GJ. Raghava GP, et al. BMC Bioinformatics. 2003 Oct 10;4:47. doi: 10.1186/1471-2105-4-47. BMC Bioinformatics. 2003. PMID: 14552658 Free PMC article. - Accuracy of structure-based sequence alignment of automatic methods.
Kim C, Lee B. Kim C, et al. BMC Bioinformatics. 2007 Sep 20;8:355. doi: 10.1186/1471-2105-8-355. BMC Bioinformatics. 2007. PMID: 17883866 Free PMC article. - Motif-Aware PRALINE: Improving the alignment of motif regions.
Dijkstra M, Bawono P, Abeln S, Feenstra KA, Fokkink W, Heringa J. Dijkstra M, et al. PLoS Comput Biol. 2018 Nov 1;14(11):e1006547. doi: 10.1371/journal.pcbi.1006547. eCollection 2018 Nov. PLoS Comput Biol. 2018. PMID: 30383764 Free PMC article. - DIALIGN-T: an improved algorithm for segment-based multiple sequence alignment.
Subramanian AR, Weyer-Menkhoff J, Kaufmann M, Morgenstern B. Subramanian AR, et al. BMC Bioinformatics. 2005 Mar 22;6:66. doi: 10.1186/1471-2105-6-66. BMC Bioinformatics. 2005. PMID: 15784139 Free PMC article.
Cited by
- Assessing the efficiency of multiple sequence alignment programs.
Pais FS, Ruy PC, Oliveira G, Coimbra RS. Pais FS, et al. Algorithms Mol Biol. 2014 Mar 6;9(1):4. doi: 10.1186/1748-7188-9-4. Algorithms Mol Biol. 2014. PMID: 24602402 Free PMC article. - The identification of short linear motif-mediated interfaces within the human interactome.
Weatheritt RJ, Luck K, Petsalaki E, Davey NE, Gibson TJ. Weatheritt RJ, et al. Bioinformatics. 2012 Apr 1;28(7):976-82. doi: 10.1093/bioinformatics/bts072. Epub 2012 Feb 10. Bioinformatics. 2012. PMID: 22328783 Free PMC article. - SLiMPrints: conservation-based discovery of functional motif fingerprints in intrinsically disordered protein regions.
Davey NE, Cowan JL, Shields DC, Gibson TJ, Coldwell MJ, Edwards RJ. Davey NE, et al. Nucleic Acids Res. 2012 Nov;40(21):10628-41. doi: 10.1093/nar/gks854. Epub 2012 Sep 12. Nucleic Acids Res. 2012. PMID: 22977176 Free PMC article. - Exact p-values for pairwise comparison of Friedman rank sums, with application to comparing classifiers.
Eisinga R, Heskes T, Pelzer B, Te Grotenhuis M. Eisinga R, et al. BMC Bioinformatics. 2017 Jan 25;18(1):68. doi: 10.1186/s12859-017-1486-2. BMC Bioinformatics. 2017. PMID: 28122501 Free PMC article. - ELM: the status of the 2010 eukaryotic linear motif resource.
Gould CM, Diella F, Via A, Puntervoll P, Gemünd C, Chabanis-Davidson S, Michael S, Sayadi A, Bryne JC, Chica C, Seiler M, Davey NE, Haslam N, Weatheritt RJ, Budd A, Hughes T, Pas J, Rychlewski L, Travé G, Aasland R, Helmer-Citterich M, Linding R, Gibson TJ. Gould CM, et al. Nucleic Acids Res. 2010 Jan;38(Database issue):D167-80. doi: 10.1093/nar/gkp1016. Epub 2009 Nov 17. Nucleic Acids Res. 2010. PMID: 19920119 Free PMC article.
References
- Mulder NJ, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Buillard V, Cerutti L, Copley R, Courcelle E, Das U, Daugherty L, Dibley M, Finn R, Fleischmann W, Gough J, Haft D, Hulo N, Hunter S, Kahn D, Kanapin A, Kejariwal A, Labarga A, Langendijk-Genevaux PS, Lonsdale D, Lopez R, Letunic I, Madera M, Maslen J, McAnulla C, McDowall J, Mistry J, Mitchell A, Nikolskaya AN, Orchard S, Orengo C, Petryszak R, Selengut JD, Sigrist CJ, Thomas PD, Valentin F, Wilson D, Wu CH, Yeats C. New developments in the InterPro database. Nucleic Acids Res. 2007;35:D224–228. doi: 10.1093/nar/gkl841. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials