The dawn of human matrilineal diversity - PubMed (original) (raw)
. 2008 May;82(5):1130-40.
doi: 10.1016/j.ajhg.2008.04.002. Epub 2008 Apr 24.
Richard Villems, Himla Soodyall, Jason Blue-Smith, Luisa Pereira, Ene Metspalu, Rosaria Scozzari, Heeran Makkan, Shay Tzur, David Comas, Jaume Bertranpetit, Lluis Quintana-Murci, Chris Tyler-Smith, R Spencer Wells, Saharon Rosset; Genographic Consortium
Collaborators, Affiliations
- PMID: 18439549
- PMCID: PMC2427203
- DOI: 10.1016/j.ajhg.2008.04.002
The dawn of human matrilineal diversity
Doron M Behar et al. Am J Hum Genet. 2008 May.
Abstract
The quest to explain demographic history during the early part of human evolution has been limited because of the scarce paleoanthropological record from the Middle Stone Age. To shed light on the structure of the mitochondrial DNA (mtDNA) phylogeny at the dawn of Homo sapiens, we constructed a matrilineal tree composed of 624 complete mtDNA genomes from sub-Saharan Hg L lineages. We paid particular attention to the Khoi and San (Khoisan) people of South Africa because they are considered to be a unique relic of hunter-gatherer lifestyle and to carry paternal and maternal lineages belonging to the deepest clades known among modern humans. Both the tree phylogeny and coalescence calculations suggest that Khoisan matrilineal ancestry diverged from the rest of the human mtDNA pool 90,000-150,000 years before present (ybp) and that at least five additional, currently extant maternal lineages existed during this period in parallel. Furthermore, we estimate that a minimum of 40 other evolutionarily successful lineages flourished in sub-Saharan Africa during the period of modern human dispersal out of Africa approximately 60,000-70,000 ybp. Only much later, at the beginning of the Late Stone Age, about 40,000 ybp, did introgression of additional lineages occur into the Khoisan mtDNA pool. This process was further accelerated during the recent Bantu expansions. Our results suggest that the early settlement of humans in Africa was already matrilineally structured and involved small, separately evolving isolated populations.
Figures
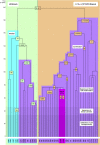
Figure 1
Simplified Human mtDNA Phylogeny The L0 and L1′5 branches are highlighted in light green and tan, respectively. The branches are made up of haplogroups L0–L6 which, in their turn, are divided into clades. Khoisan and non-Khoisan clades are shown in blue and purple, respectively. Clades involved in the African exodus are shown in pink. A time scale is given on the left. Approximate time periods for the beginning of African LSA modernization, appearance of African LSA sites, and solidization of LSA throughout Africa are shown by increasing colors densities. For a more detailed phylogeny, see Figure S1.
Figure 2
Maternal Gene Flow within Africa The gradual maternal movements suggested by the first (A) and second (B) hypotheses are denoted by the ascending numerical labels. A gradient colorization system is used to illustrate the timing of the events. The temporal direction and timing of the arrows and expansion waves are general and should not be treated as firm migratory paths. (A) An initial prolonged colonization (brown) by anatomically modern humans (1) is followed by a dispersal wave (green) of a fracture of the population (2) and the localization of L0d and L0k to southern Africa (3). (B) An early Homo sapiens division in a hypothetical migration zone (1) resulted in two separately evolving populations (2) and the localization of L0 (green) in southern Africa and L1′5 (red) in eastern Africa. A subsequent dispersal event of the L0abf subset from the southern population and its mergence with the eastern population (brown) is suggested (3), resulting in the former population composed only of L0d and L0k and the latter composed of L1′5 and L0abf. Later dispersal waves from the eastern African population parallels the beginning of African LSA approximately 70,000 ybp (4). Rapid migrations during the LSA (5) brought descendants of the eastern African population into repeated contact with the southern population, peaking during the Bantu expansion (6).
Similar articles
- Whole-mtDNA genome sequence analysis of ancient African lineages.
Gonder MK, Mortensen HM, Reed FA, de Sousa A, Tishkoff SA. Gonder MK, et al. Mol Biol Evol. 2007 Mar;24(3):757-68. doi: 10.1093/molbev/msl209. Epub 2006 Dec 28. Mol Biol Evol. 2007. PMID: 17194802 - Ancient substructure in early mtDNA lineages of southern Africa.
Barbieri C, Vicente M, Rocha J, Mpoloka SW, Stoneking M, Pakendorf B. Barbieri C, et al. Am J Hum Genet. 2013 Feb 7;92(2):285-92. doi: 10.1016/j.ajhg.2012.12.010. Epub 2013 Jan 17. Am J Hum Genet. 2013. PMID: 23332919 Free PMC article. - On the edge of Bantu expansions: mtDNA, Y chromosome and lactase persistence genetic variation in southwestern Angola.
Coelho M, Sequeira F, Luiselli D, Beleza S, Rocha J. Coelho M, et al. BMC Evol Biol. 2009 Apr 21;9:80. doi: 10.1186/1471-2148-9-80. BMC Evol Biol. 2009. PMID: 19383166 Free PMC article. - The reversal of human phylogeny: Homo left Africa as erectus, came back as sapiens sapiens.
Árnason Ú, Hallström B. Árnason Ú, et al. Hereditas. 2020 Dec 19;157(1):51. doi: 10.1186/s41065-020-00163-9. Hereditas. 2020. PMID: 33341120 Free PMC article. Review. - African human mtDNA phylogeography at-a-glance.
Rosa A, Brehem A. Rosa A, et al. J Anthropol Sci. 2011;89:25-58. doi: 10.4436/jass.89006. Epub 2011 Mar 15. J Anthropol Sci. 2011. PMID: 21368343 Review.
Cited by
- A worldwide survey of human male demographic history based on Y-SNP and Y-STR data from the HGDP-CEPH populations.
Shi W, Ayub Q, Vermeulen M, Shao RG, Zuniga S, van der Gaag K, de Knijff P, Kayser M, Xue Y, Tyler-Smith C. Shi W, et al. Mol Biol Evol. 2010 Feb;27(2):385-93. doi: 10.1093/molbev/msp243. Epub 2009 Oct 12. Mol Biol Evol. 2010. PMID: 19822636 Free PMC article. - The trans-Saharan slave trade - clues from interpolation analyses and high-resolution characterization of mitochondrial DNA lineages.
Harich N, Costa MD, Fernandes V, Kandil M, Pereira JB, Silva NM, Pereira L. Harich N, et al. BMC Evol Biol. 2010 May 10;10:138. doi: 10.1186/1471-2148-10-138. BMC Evol Biol. 2010. PMID: 20459715 Free PMC article. - New insights into the Lake Chad Basin population structure revealed by high-throughput genotyping of mitochondrial DNA coding SNPs.
Cerezo M, Černý V, Carracedo Á, Salas A. Cerezo M, et al. PLoS One. 2011 Apr 20;6(4):e18682. doi: 10.1371/journal.pone.0018682. PLoS One. 2011. PMID: 21533064 Free PMC article. - The case for the continuing use of the revised Cambridge Reference Sequence (rCRS) and the standardization of notation in human mitochondrial DNA studies.
Bandelt HJ, Kloss-Brandstätter A, Richards MB, Yao YG, Logan I. Bandelt HJ, et al. J Hum Genet. 2014 Feb;59(2):66-77. doi: 10.1038/jhg.2013.120. Epub 2013 Dec 5. J Hum Genet. 2014. PMID: 24304692 Review. - Genetic portrait of Lisboa immigrant population from Cabo Verde with mitochondrial DNA analysis.
Morais P, Amorim A, Vieira da Silva C, Ribeiro T, Costa Santos J, Afonso Costa H. Morais P, et al. J Genet. 2015 Sep;94(3):509-12. doi: 10.1007/s12041-015-0552-7. J Genet. 2015. PMID: 26440093 No abstract available.
References
- Cann R.L., Stoneking M., Wilson A.C. Mitochondrial DNA and human evolution. Nature. 1987;325:31–36. - PubMed
- Underhill P.A., Kivisild T. Use of Y chromosome and mitochondrial DNA population structure in tracing human migrations. Annu. Rev. Genet. 2007;41:539–564. - PubMed
- Mellars P. Going east: New genetic and archaeological perspectives on the modern human colonization of Eurasia. Science. 2006;313:796–800. - PubMed
- Macaulay V., Hill C., Achilli A., Rengo C., Clarke D., Meehan W., Blackburn J., Semino O., Scozzari R., Cruciani F. Single, rapid coastal settlement of Asia revealed by analysis of complete mitochondrial genomes. Science. 2005;308:1034–1036. - PubMed
- Torroni A., Achilli A., Macaulay V., Richards M., Bandelt H.J. Harvesting the fruit of the human mtDNA tree. Trends Genet. 2006;22:339–345. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources