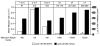Division of labor at the eukaryotic replication fork - PubMed (original) (raw)
Division of labor at the eukaryotic replication fork
Stephanie A Nick McElhinny et al. Mol Cell. 2008.
Abstract
DNA polymerase delta (Pol delta) and DNA polymerase epsilon (Pol epsilon) are both required for efficient replication of the nuclear genome, yet the division of labor between these enzymes has remained unclear for many years. Here we investigate the contribution of Pol delta to replication of the leading and lagging strand templates in Saccharomyces cerevisiae using a mutant Pol delta allele (pol3-L612M) whose error rate is higher for one mismatch (e.g., T x dGTP) than for its complement (A x dCTP). We find that strand-specific mutation rates strongly depend on the orientation of a reporter gene relative to an adjacent replication origin, in a manner implying that >90% of Pol delta replication is performed using the lagging strand template. When combined with recent evidence implicating Pol epsilon in leading strand replication, these data support a model of the replication fork wherein the leading and lagging strand templates are primarily copied by Pol epsilon and Pol delta, respectively.
Figures
Figure 1. Preferential Replication of the Lagging Strand Template by L612M Pol δ
The orientation (5′ to 3′ with respect to coding sequence) of the URA3 reporter (gray arrow) is depicted by the direction of the arrow. URA3 hotspot locations are identified numerically within the arrow. (A) URA3 orientation 1 (OR1); (B) URA3 orientation 2 (OR2). The lagging (green) and leading (blue) strand templates of the six mutational hotspots are shown, with the hotspot sequence of the preferred templating strand (based on the error bias of L612M Pol δ) shown in bold black font. The observed errors at each site (shown in red above or below the template sequence) are distributed between the leading and lagging strand templates according to the biased error rates of L612M Pol δ determined in vitro using the lacZ forward mutation assay. For the three base substitution hotspots, the mismatch that causes the lagging strand errors is shown in parentheses with the templating base in bold black font and the mismatched incoming nucleotide in red. Mutation rates for lagging strand (green values) and leading strand (blue values) errors for each hotspot were calculated by dividing the number of events assigned to each strand by the total number of events observed in that orientation and then multiplying by the ura3 forward mutation rate of the strain.
Figure 2. Orientation Dependence of L612M Pol δ Mutagenesis in the URA3 Target
The mutation rate in orientation 1 (OR1, upward black bars) and orientation 2 (OR2, downward gray bars) for each type of error is shown for the hotspot locations (numerical values) and all other sites within the URA3 target (other). When an event was not observed, the upper limit of the mutation rate is shown, labeled with a ≤ symbol above (OR1) or below (OR2) the bar. See Table S1 for mutation rate values.
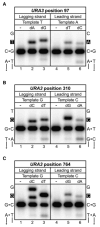
Figure 3. Error Bias of L612M Pol δ at the Three URA3 Base Substitution Hotspots
Mismatch insertion efficiency of L612M Pol δ was determined in vitro for lagging versus leading strand errors for the URA3 base substitution hotspots at positions (A) 97, (B) 310, and (C) 764. For each panel, the template sequences of the lagging and leading strands are shown on the left and right, respectively, with the site of insertion in white font on a black background. Lanes 1 and 4 of each panel are mock reactions lacking L612M Pol δ; lanes 2 and 5 contain the correct incoming nucleotide (100 μM, 1 min reaction); and lanes 3 and 6 contain the appropriate mismatched nucleotide (1 mM, 10 min reaction). See Table S2 for additional information.
Figure 4. Mismatch Repair Efficiency of the L612M Pol δ Hotspot Mutations
The mutation rate for each hotspot mutation (in the orientation in which it was hot) is shown for mismatch repair-proficient (pol3-L612M MSH2, white bars, scale on left axis) and deficient (_pol3-L612M msh2_Δ, black bars, scale on right axis in white font on black background) strains. The observed mutation and URA3 position are shown above each set of bars. The mismatch repair factor (shown below each hotspot mutation) is the ratio of the mutation rates for the two strains. When an event was not observed, the upper limit of the mutation rate is shown, labeled with a ≤ symbol above the bar. See Tables S1 and S3 for mutation rate values.
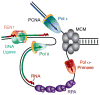
Figure 5. A Simple Model for the Division of Labor at the Eukaryotic Replication Fork
In this model, replication of the leading and lagging strands is performed by Pol ε and Pol δ, respectively. Only the MCM helicase complex is shown at the fork junction, but many other proteins participate in this process. The minimal Okazaki fragment maturation machinery on the lagging strand is shown (see Garg and Burgers, 2005).
Comment in
- DNA polymerases at the replication fork in eukaryotes.
Stillman B. Stillman B. Mol Cell. 2008 May 9;30(3):259-60. doi: 10.1016/j.molcel.2008.04.011. Mol Cell. 2008. PMID: 18471969 Free PMC article.
References
- Burgers PM, Gerik KJ. Structure and processivity of two forms of Saccharomyces cerevisiae DNA polymerase delta. J Biol Chem. 1998;273:19756–19762. - PubMed
- Buschiazzo E, Gemmell NJ. The rise, fall and renaissance of microsatellites in eukaryotic genomes. Bioessays. 2006;28:1040–1050. - PubMed
- Dua R, Levy DL, Campbell JL. Role of the putative zinc finger domain of Saccharomyces cerevisiae DNA polymerase epsilon in DNA replication and the S/M checkpoint pathway. J Biol Chem. 1998;273:30046–30055. - PubMed
- Dua R, Levy DL, Campbell JL. Analysis of the essential functions of the C-terminal protein/protein interaction domain of Saccharomyces cerevisiae Pol epsilon and its unexpected ability to support growth in the absence of the DNA polymerase domain. J Biol Chem. 1999;274:22283–22288. - PubMed
- Eshleman JR, Markowitz SD. Microsatellite instability in inherited and sporadic neoplasms. Curr Opin Oncol. 1995;7:83–89. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases