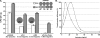Tools for functional postgenomic analysis of listeria monocytogenes - PubMed (original) (raw)
Tools for functional postgenomic analysis of listeria monocytogenes
Ian R Monk et al. Appl Environ Microbiol. 2008 Jul.
Abstract
We describe the development of genetic tools for regulated gene expression, the introduction of chromosomal mutations, and improved plasmid transfer by electroporation in the food-borne pathogen Listeria monocytogenes. pIMK, a kanamycin-resistant, site-specific, integrative listeriophage vector was constructed and then modified for overexpression (pIMK2) or for isopropyl-beta-d-thiogalactopyranoside (IPTG)-regulated expression (pIMK3 and pIMK4). The dynamic range of promoters was assessed by determining luciferase activity, P60 secretion, and internalin A-mediated invasion. These analyses demonstrated that pIMK4 and pIMK3 have a stringently controlled dynamic range of 540-fold. Stable gene overexpression was achieved with pIMK2, giving a range of expression for the three vectors of 1,350-fold. The lactococcal pORI280 system was optimized for the generation of chromosomal mutations and used to create five new prfA star mutants. The combination of pIMK4 and pORI280 allowed streamlined creation of "IPTG-dependent" mutants. This was exemplified by creation of a clean deletion mutant with deletion of the universally essential secA gene, and this mutant exhibited a rapid loss of viability upon withdrawal of IPTG. We also improved plasmid transfer by electroporation into three commonly used laboratory strains of L. monocytogenes. A 125-fold increase in transformation efficiency for EGDe compared with the widely used protocol of Park and Stewart (S. F. Park and G. S. Stewart, Gene 94:129-132, 1990) was observed. Maximal transformation efficiencies of 5.7 x 10(6) and 6.7 x 10(6) CFU per mug were achieved for EGDe and 10403S, respectively, with a replicating plasmid. An efficiency of 2 x 10(7) CFU per mug is the highest efficiency reported thus far for L. monocytogenes F2365.
Figures
FIG. 1.
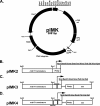
Plasmid maps of pIMK and promoter derivatives. (A) Plasmid pIMK was created by SOE PCR as described in Materials and Methods. This plasmid is based on the pPL2 vector (37), which replicates autonomously in E. coli but is unable to replicate in L. monocytogenes. Integration into the tRNAArg locus is directed by the PSA phage integrase under the control of the listerial P60 promoter and confers kanamycin resistance. For sequencing purposes, T3 and T7 primer binding sites are present before the KpnI restriction site and after the SacI restriction site, respectively. The restriction sites labeled on pIMK are unique. (B to D) Derivatives of pIMK were constructed to enable heterologous gene expression through in-frame cloning of promoterless genes via a unique NcoI site overlapping the start codon. Additional restriction sites used for cloning are underlined. (B) pIMK2 enables constitutive overexpression of genes from the synthetic Phelp promoter (59). (C) pIMK3. There is high-level, IPTG-controlled expression from the Phelp promoter. A consensus lacOid sequence (LacI binding site [gray box] [51]) was inserted between the PCP25 promoter (containing the −35 and −10 regions) and the downstream 5′ untranslated region of the hlyA gene. (D) pIMK4. There is strict IPTG-controlled expression from the pIMK3 promoter with an additional lacOid sequence (gray box) incorporated directly upstream of the PCP25 region. In both pIMK3 and pIMK4, the lacI repressor is expressed from the Phelp promoter, which is cloned between the SalI and KpnI restriction sites.
FIG. 2.
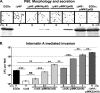
Expression profiles of pIMK4, pIMK3 (IPTG inducible), and pIMK2 (overexpression) demonstrated by P60 secretion and InlA-mediated invasion. (A) Gram staining (top panel) and anti-P60-specific Western blotting (bottom panel) for the secreted proteins from BHI-grown, exponential-phase (OD600, 1.0) cells transformed with the pIMK derivatives containing p60. (B) Gentamicin protection assay for InlA-mediated invasion of Caco-2 cells. Strains transformed with the pIMK derivatives containing inlA were grown to exponential phase (OD600,1.0) in BHI containing various concentrations of IPTG and added to Caco-2 cells as described in Materials and Methods. Invasion was expressed as the total number of CFU recovered per well, and each mean and standard deviation are the results of one representative experiment performed in quadruplicate. Inclusion of IPTG in the preculture did not impact the invasion by EGDe. Statistical analyses were performed using the raw CFU counts and the Student t test, and P values less than 0.01 were considered significant (**, P < 0.005; ***, P < 0.001). NS indicates that the P value is above the level of significance.
FIG. 3.
Improved electroporation protocol for L. monocytogenes. Systematic changes to the protocol of Park and Stewart (53) (P&S) led to the development of the improved electroporation protocol (NEW) as described in Materials and Methods. A series of replicative (pNZ8048) and integrative (pPL2, pIMK, and pPL2_lux_-Phelp) plasmids were tested to compare the efficiency of the protocol of Park and Stewart and the efficiency of the new protocol. Most transformants were selected on BHI agar containing chloramphenicol at a concentration of 7.5 μg/ml; the only exception was pIMK, which was selected on medium containing kanamycin at a concentration of 50 μg/ml. The images in the top right of panel A show 10-μl spot dilutions (10−1, 10−2, 10−3, and 10−4) from pNZ8048 transformations for the protocol of Park and Stewart and the new protocol. Colonies obtained with the new protocol recovered faster than colonies obtained with the protocol of Park and Stewart, as shown by larger colonies. The images of plates (transformed with the new protocol) from pPL2 and pIMK transformations show the uniformity of the colony size for pIMK-containing cells compared to the pPL2-containing cells. Colonies were visible after overnight incubation of pIMK-transformed strains. Statistical analyses were preformed using the raw CFU counts and the Student t test, and P values less than 0.01 were considered significant (***, P < 0.001). (B) Relative contributions of lysozyme treatment for EGDe (circles) and F2365 (triangles) expressed as a percentage of the maximal transformation. The highest levels of transformation were obtained with 10 and 25 μg/ml lysozyme for EGDe and F2365, respectively.
FIG. 4.
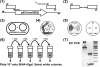
Rapid gene deletion protocol for L. monocytogenes: the pORI280 system (41). The following steps are shown diagrammatically. In step 1, an SOE PCR product was generated with 400-bp upstream (AB to the start codon) and downstream (CD from the stop codon) fragments of the gene to be deleted. In step 2, the AB and CD fragments were joined by a second round of PCR with the A and D primers to form the AD product. This amplimer was cloned into pORI280, generating pORI280(AD). In step 3, pORI280(AD) and the RepA-supplying temperature-sensitive plasmid pVE6007 were cotransformed into the recipient electrocompetent L. monocytogenes strain and selected on BHI agar containing 5 μg/ml erythromycin and 100 μg/ml X-Gal at 30°C for 48 h. In step 4, single blue colonies were complex streaked on BHI agar containing erythromycin and X-Gal and then incubated at 37°C for 24 h. This step resulted in the loss of pVE6007 and then caused pORI280AD to integrate via the AB or CD crossover. In step 5, light and dark blue colonies arising from pORI280AD integration were patched onto both BHI agar containing 5 μg/ml erythromycin and BHI agar containing 7.5 μg/ml chloramphenicol (Cm) and then incubated at 37°C for 24 h. NG indicates no growth on chloramphenicol plates and loss of pVE6007. In step 6, a single light blue colony and a single dark blue colony were grown statically to stationary phase at 37°C. The cultures were diluted 1:1,000 in fresh BHI, and the process was repeated for five sequential passages. Each of the five passages was diluted 10−5, 100 μl was spread plated onto BHI agar containing X-Gal, and the plates were incubated at 37°C for 24 h. In step 7, white colonies were screened by colony PCR with the E and F primers as shown for step 1. Colonies were also tested for erythromycin sensitivity by patching for the loss of pORI280(AD), and the EF amplimer was sequenced. Wt, wild type.
FIG. 5.
Site-directed chromosomal mutagenesis using pORI280: new prfA star mutants. The procedure described in the legend to Fig. 4 was used to create single nucleotide point mutations in the chromosome. In steps 1 and 2, SOE PCR was used to create directed nucleotide changes in a region of DNA, and both the B and C primers contained the desired mutations (indicated by three stars). However, in this case, genomic DNA from naturally induced prfA mutants was used as the template. PrfA mutants were generated from a hemolysin promoter fused to a chloramphenicol reporter gene (EGDe::p_hly_C), with selection pressure (BHI agar containing 50 μg/ml of chloramphenicol) to induce prfA mutations. Mutants were recreated using the pORI280 system in a fresh EGDe background. Five microliters of an overnight BHI culture of each recreated isolate was spotted onto BHI agar containing (i) 5% defibrinated sheep blood with 1 U/100 ml sphingomyelinase (Sigma), (ii) 4% lecithin (Oxoid) (PCPLC), (iii) 0.2%
l
-α-phosphatidylinositol (1 g dissolved in 25 ml distilled water, autoclaved, and added to 475 ml BHI agar at 50°C) (Sigma) (PIPLC), and (iv) 0.2% freeze-dried Micrococcus luteus cells (cell wall hydrolase activity) (Sigma). Plates were incubated for 48 h at 37°C. Wt, wild type.
FIG. 6.
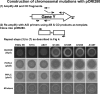
Essential gene deletion in L. monocytogenes: secA is an essential gene. The pORI system was combined with the strict IPTG-inducible promoter (pIMK4) for inactivation of an essential gene. The procedure is outlined in panel A for deletion of the essential preprotein translocase gene secA. (B) An overnight culture of EGDeΔ_secA_::pIMK4_secA_ (grown in BHI containing 1 mM IPTG) was washed three times with BHI and diluted 1:100 in fresh medium with (filled circles) or without (open circles) 1 mM IPTG. Cultures were grown at 37°C with shaking, and samples were taken at the indicated time points. Cells were then diluted in PBS, plated onto BHI agar containing 1 mM IPTG, and incubated overnight before colony enumeration. EGDeΔ_secA_::pIMK4_secA_ exhibited a wild-type growth profile (data not shown). (C) An overnight culture of EGDeΔ_secA_::pIMK4_secA_ (grown in BHI containing 1 mM IPTG) was diluted 10−5 and plated onto BHI agar containing different concentrations of IPTG, as indicated. The plates were incubated at 37°C for 24 h before visualization.
Similar articles
- Generation of nonpolar deletion mutants in Listeria monocytogenes using the "SOEing" method.
Rychli K, Guinane CM, Daly K, Hill C, Cotter PD. Rychli K, et al. Methods Mol Biol. 2014;1157:187-200. doi: 10.1007/978-1-4939-0703-8_16. Methods Mol Biol. 2014. PMID: 24792559 - Generation of Nonpolar Deletion Mutants in Listeria monocytogenes Using the "SOEing" Method.
Rychli K, Wagner E, Guinane CM, Daly K, Hill C, Cotter PD. Rychli K, et al. Methods Mol Biol. 2021;2220:165-175. doi: 10.1007/978-1-0716-0982-8_13. Methods Mol Biol. 2021. PMID: 32975774 - Construction, characterization, and use of two Listeria monocytogenes site-specific phage integration vectors.
Lauer P, Chow MY, Loessner MJ, Portnoy DA, Calendar R. Lauer P, et al. J Bacteriol. 2002 Aug;184(15):4177-86. doi: 10.1128/JB.184.15.4177-4186.2002. J Bacteriol. 2002. PMID: 12107135 Free PMC article. - The Listeria monocytogenes iap gene as an indicator gene for the study of PrfA-dependent regulation.
Bubert A, Kestler H, Götz M, Böckmann R, Goebel W. Bubert A, et al. Mol Gen Genet. 1997 Sep;256(1):54-62. doi: 10.1007/s004380050545. Mol Gen Genet. 1997. PMID: 9341679 - Identification and characterization of a novel PrfA-regulated gene in Listeria monocytogenes whose product, IrpA, is highly homologous to internalin proteins, which contain leucine-rich repeats.
Domann E, Zechel S, Lingnau A, Hain T, Darji A, Nichterlein T, Wehland J, Chakraborty T. Domann E, et al. Infect Immun. 1997 Jan;65(1):101-9. doi: 10.1128/iai.65.1.101-109.1997. Infect Immun. 1997. PMID: 8975898 Free PMC article.
Cited by
- Aquatic environment drives the emergence of cell wall-deficient dormant forms in Listeria.
Carvalho F, Carreaux A, Sartori-Rupp A, Tachon S, Gazi AD, Courtin P, Nicolas P, Dubois-Brissonnet F, Barbotin A, Desgranges E, Bertrand M, Gloux K, Schouler C, Carballido-López R, Chapot-Chartier MP, Milohanic E, Bierne H, Pagliuso A. Carvalho F, et al. Nat Commun. 2024 Oct 2;15(1):8499. doi: 10.1038/s41467-024-52633-7. Nat Commun. 2024. PMID: 39358320 Free PMC article. - Maple compounds prevent biofilm formation in Listeria monocytogenes via sortase inhibition.
Elbakush AM, Trunschke O, Shafeeq S, Römling U, Gomelsky M. Elbakush AM, et al. Front Microbiol. 2024 Sep 16;15:1436476. doi: 10.3389/fmicb.2024.1436476. eCollection 2024. Front Microbiol. 2024. PMID: 39351304 Free PMC article. - EⅡB Mutation Reduces the Pathogenicity of Listeria monocytogenes by Negatively Regulating Biofilm Formation Ability, Infective Capacity, and Virulence Gene Expression.
Liu C, Qian R, Shi W, Kou L, Wang J, Ma X, Ren H, Gao S, Ren J. Liu C, et al. Vet Sci. 2024 Jul 2;11(7):301. doi: 10.3390/vetsci11070301. Vet Sci. 2024. PMID: 39057985 Free PMC article. - An insight into the role of branched-chain α-keto acid dehydrogenase (BKD) complex in branched-chain fatty acid biosynthesis and virulence of Listeria monocytogenes.
Kader Chowdhury QMM, Islam S, Narayanan L, Ogunleye SC, Wang S, Thu D, Freitag NE, Lawrence ML, Abdelhamed H. Kader Chowdhury QMM, et al. J Bacteriol. 2024 Jul 25;206(7):e0003324. doi: 10.1128/jb.00033-24. Epub 2024 Jun 20. J Bacteriol. 2024. PMID: 38899896 - Role of ethanolamine utilization and bacterial microcompartment formation in Listeria monocytogenes intracellular infection.
Chatterjee A, Kaval KG, Garsin DA. Chatterjee A, et al. Infect Immun. 2024 Jun 11;92(6):e0016224. doi: 10.1128/iai.00162-24. Epub 2024 May 16. Infect Immun. 2024. PMID: 38752742 Free PMC article.
References
- Alberti-Segui, C., K. R. Goeden, and D. E. Higgins. 2007. Differential function of Listeria monocytogenes listeriolysin O and phospholipases C in vacuolar dissolution following cell-to-cell spread. Cell. Microbiol. 9:179-195. - PubMed
- Archambaud, C., M. A. Nahori, J. Pizarro-Cerda, P. Cossart, and O. Dussurget. 2006. Control of Listeria superoxide dismutase by phosphorylation. J. Biol. Chem. 281:31812-31822. - PubMed
- Begley, M., C. Hill, and C. G. Gahan. 2003. Identification and disruption of btlA, a locus involved in bile tolerance and general stress resistance in Listeria monocytogenes. FEMS Microbiol. Lett. 218:31-38. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources