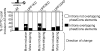Regulation of multiple core spliceosomal proteins by alternative splicing-coupled nonsense-mediated mRNA decay - PubMed (original) (raw)
Regulation of multiple core spliceosomal proteins by alternative splicing-coupled nonsense-mediated mRNA decay
Arneet L Saltzman et al. Mol Cell Biol. 2008 Jul.
Abstract
Alternative splicing (AS) can regulate gene expression by introducing premature termination codons (PTCs) into spliced mRNA that subsequently elicit transcript degradation by the nonsense-mediated mRNA decay (NMD) pathway. However, the range of cellular functions controlled by this process and the factors required are poorly understood. By quantitative AS microarray profiling, we find that there are significant overlaps among the sets of PTC-introducing AS events affected by individual knockdown of the three core human NMD factors, Up-Frameshift 1 (UPF1), UPF2, and UPF3X/B. However, the levels of some PTC-containing splice variants are less or not detectably affected by the knockdown of UPF2 and/or UPF3X, compared with the knockdown of UPF1. The intron sequences flanking the affected alternative exons are often highly conserved, suggesting important regulatory roles for these AS events. The corresponding genes represent diverse cellular functions, and surprisingly, many encode core spliceosomal proteins and assembly factors. We further show that conserved, PTC-introducing AS events are enriched in genes that encode core spliceosomal proteins. Where tested, altering the expression levels of these core spliceosomal components affects the regulation of PTC-containing splice variants from the corresponding genes. Together, our results show that AS-coupled NMD can have different UPF factor requirements and is likely to regulate many general components of the spliceosome. The results further implicate general spliceosomal components in AS regulation.
Figures
FIG. 1.
Overlapping but distinct effects of UPF protein knockdowns on PTC-introducing AS events. (A) Western blot assays of HeLa cell lysates following siRNA-mediated UPF factor knockdowns. Western blots were probed with UPF2- or UPF3-specific antibodies (upper panels) and with calnexin- or β-actin-specific antibodies as loading controls (lower panels). Serial threefold dilutions of control lysates are shown in the left panels. An estimation of the knockdown efficiency following targeting siRNA treatment relative to the control siRNA treatment is shown below the gels (right panels). For a Western blot assay showing knockdown of UPF1, see reference . (B) The change in alternative exon inclusion level (percent skipping) is shown for AS events that introduce a PTC upon inclusion (left panel) or upon skipping (right panel). The change in percent skipping is represented by the cyan-black-yellow color scale shown and is calculated for each AS event as the percent skipping in the UPF1, UPF2, or UPF3X knockdown minus the percent skipping for the same AS event in the corresponding control siRNA treatment. Events for which we detected at least a 5% change in percent skipping upon UPF1 knockdown are shown, and rows are ordered according to the median of the percent skipping change across the three knockdowns.
FIG. 2.
Representative RT-PCR assays showing effects of UPF protein knockdown on levels of PTC-introducing alternative exons. RT-PCR assays were performed with primers specific for the constitutive exons flanking the alternative exons. The RNA samples analyzed are indicated above the lanes and correspond to cells transfected with either a control siRNA (−) or an siRNA specific for the indicated UPF factor (+). Arrows to the right of each gel indicate the expected sizes of included and skipped products. The color bar below each gel panel shows the quantification of the knockdown-dependent changes in percent skipping predicted by the microarray (upper color) and measured by RT-PCR (lower color). The change in percent skipping is shown with the same color scale as in Fig. 1B. For details on these AS events, see Table S2 in the supplemental material (PTC-upon-inclusion AS events 3, 146, 957, 557, 2315, and 2372; PTC-upon-skipping AS events 750, 1268, 76, 188, 1909, and 2914).
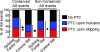
FIG. 3.
PTC-upon-inclusion alternative exons that show UPF1- or UPF2-dependent changes (at least a 5% difference) in inclusion level are significantly often flanked by highly conserved intronic sequences (*1, P = 2·10−4; *2, P = 3·10−2, Fisher's exact test; compare the proportions marked by asterisks to the proportion for the total group [All]). The stacked bar graph shows the proportion of PTC-upon-inclusion AS events that overlap (black) or do not overlap (white) phylogenetically conserved sequences, as identified by the phastCons algorithm (49). Overlap requires that at least 35 of the first 50 nucleotides of both the upstream and downstream intron sequences flanking the alternative exon overlap phastCons elements. “All” represents all detectable PTC-upon-inclusion events (n = 164), and “KD” represents PTC-upon-inclusion events with at least a 5% difference in the indicated direction (more inclusion or more skipping) and knockdown (UPF1KD more inclusion, n = 46; UPF2KD more inclusion, n = 16).
FIG. 4.
Conserved AS events in genes for spliceosomal proteins are more often PTC introducing than are those in genes from the control set (compare proportions marked by asterisks; P = 3·10−3, chi-square test). Conserved AS events either have flanking introns overlapping (≥35 nucleotides) phastCons conserved elements or are conserved in mouse based on an analysis of cDNAs/ESTs. PTC-introducing AS events in the spliceosomal protein set are also more often conserved than nonconserved (compare proportions marked by closed circles). Genes with more than one AS event of the same type (PTC introducing or non-PTC introducing) were only counted once.
FIG. 5.
SNRPB (also known as SmB/B′) or SMNDC1 (also known as SPF30) overexpression leads to increased levels of the respective PTC-containing (PTC+) alternative transcript. Either Flag-tagged SNRPB or SMNDC1 was transiently overexpressed in HeLa cells, and the endogenous PTC-containing transcript of SNRPB (A, left top panel) or SMNDC1 (A, right top panel) was amplified by RT-PCR with a forward primer specific for the 5′ untranslated region or an upstream exon and a reverse primer specific for the PTC-introducing alternative exon. RT-PCR amplification of conserved, PTC-containing transcripts of SF1, TRA2A, and SR140 were not affected to the same degree by the overexpression of SNRPB (A, left lower panels) or SMNDC1 (A, right lower panels). The overexpression of another Flag-tagged spliceosome-associated protein (DDX42) in parallel had little effect on the level of the SNRPB and SMNDC1 transcripts. (B) Western blot assay showing the expression of each Flag-tagged protein (left, approximate molecular sizes of markers in kilodaltons; right, arrowheads indicate expected protein sizes). Data are representative of three independent transfections, and RT-PCR assays were performed in triplicate.
Similar articles
- Comparison of EJC-enhanced and EJC-independent NMD in human cells reveals two partially redundant degradation pathways.
Metze S, Herzog VA, Ruepp MD, Mühlemann O. Metze S, et al. RNA. 2013 Oct;19(10):1432-48. doi: 10.1261/rna.038893.113. Epub 2013 Aug 20. RNA. 2013. PMID: 23962664 Free PMC article. - Mechanism of Nonsense-Mediated mRNA Decay Stimulation by Splicing Factor SRSF1.
Aznarez I, Nomakuchi TT, Tetenbaum-Novatt J, Rahman MA, Fregoso O, Rees H, Krainer AR. Aznarez I, et al. Cell Rep. 2018 May 15;23(7):2186-2198. doi: 10.1016/j.celrep.2018.04.039. Cell Rep. 2018. PMID: 29768215 Free PMC article. - Quantitative microarray profiling provides evidence against widespread coupling of alternative splicing with nonsense-mediated mRNA decay to control gene expression.
Pan Q, Saltzman AL, Kim YK, Misquitta C, Shai O, Maquat LE, Frey BJ, Blencowe BJ. Pan Q, et al. Genes Dev. 2006 Jan 15;20(2):153-8. doi: 10.1101/gad.1382806. Genes Dev. 2006. PMID: 16418482 Free PMC article. - Upf proteins: highly conserved factors involved in nonsense mRNA mediated decay.
Gupta P, Li YR. Gupta P, et al. Mol Biol Rep. 2018 Feb;45(1):39-55. doi: 10.1007/s11033-017-4139-7. Epub 2017 Dec 27. Mol Biol Rep. 2018. PMID: 29282598 Review. - UPF1 P-body localization.
Brogna S, Ramanathan P, Wen J. Brogna S, et al. Biochem Soc Trans. 2008 Aug;36(Pt 4):698-700. doi: 10.1042/BST0360698. Biochem Soc Trans. 2008. PMID: 18631143 Review.
Cited by
- An autoregulatory poison exon in Smndc1 is conserved across kingdoms and influences organism growth.
Belleville AE, Thomas JD, Tonnies J, Gabel AM, Borrero Rossi A, Singh P, Queitsch C, Bradley RK. Belleville AE, et al. PLoS Genet. 2024 Aug 16;20(8):e1011363. doi: 10.1371/journal.pgen.1011363. eCollection 2024 Aug. PLoS Genet. 2024. PMID: 39150991 Free PMC article. - Functional investigation of SCN1A deep-intronic variants activating poison exons inclusion.
Sparber P, Bychkov I, Pyankov D, Skoblov M. Sparber P, et al. Hum Genet. 2023 Aug;142(8):1043-1053. doi: 10.1007/s00439-023-02564-y. Epub 2023 Apr 25. Hum Genet. 2023. PMID: 37186029 - The oncogenic role of SNRPB in human tumors: A pan-cancer analysis.
Wu J, Lu F, Yu B, Wang W, Ye X. Wu J, et al. Front Mol Biosci. 2022 Oct 6;9:994440. doi: 10.3389/fmolb.2022.994440. eCollection 2022. Front Mol Biosci. 2022. PMID: 36275630 Free PMC article. - Non-ubiquitous expression of core spliceosomal protein SmB/B' in chick and mouse embryos.
Turner BRH, Mellor C, McElroy C, Bowen N, Gu W, Knill C, Itasaki N. Turner BRH, et al. Dev Dyn. 2023 Feb;252(2):276-293. doi: 10.1002/dvdy.537. Epub 2022 Sep 20. Dev Dyn. 2023. PMID: 36058892 Free PMC article. - Global SLAM-seq for accurate mRNA decay determination and identification of NMD targets.
Alalam H, Zepeda-Martínez JA, Sunnerhagen P. Alalam H, et al. RNA. 2022 Jun;28(6):905-915. doi: 10.1261/rna.079077.121. Epub 2022 Mar 16. RNA. 2022. PMID: 35296539 Free PMC article.
References
- Amrani, N., M. S. Sachs, and A. Jacobson. 2006. Early nonsense: mRNA decay solves a translational problem. Nat. Rev. Mol. Cell Biol. 7415-425. - PubMed
- Black, D. L. 2003. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 72291-336. - PubMed
- Blencowe, B. J. 2006. Alternative splicing: new insights from global analyses. Cell 12637-47. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials