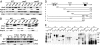Nuclear translocation of Gln3 in response to nutrient signals requires Golgi-to-endosome trafficking in Saccharomyces cerevisiae - PubMed (original) (raw)
Nuclear translocation of Gln3 in response to nutrient signals requires Golgi-to-endosome trafficking in Saccharomyces cerevisiae
Rekha Puria et al. Proc Natl Acad Sci U S A. 2008.
Abstract
The yeast Saccharomyces cerevisiae has developed specialized mechanisms that enable growth on suboptimal nitrogen sources. Exposure of yeast cells to poor nitrogen sources or treatment with the Tor kinase inhibitor rapamycin elicits activation of Gln3 and transcription of nitrogen catabolite-repressed (NCR) genes whose products function in scavenging and metabolizing nitrogen. Here, we show that mutations in class C and D Vps components, which mediate Golgi-to-endosome vesicle transport, impair nuclear translocation of Gln3, NCR gene activation, and growth in poor nitrogen sources. In nutrient-replete conditions, a significant fraction of Gln3 is peripherally associated with light membranes and partially colocalizes with Vps10-containing foci. These results reveal a role for Golgi-to-endosome vesicular trafficking in TORC1-controlled nuclear translocation of Gln3 and support a model in which Tor-mediated signaling in response to nutrient cues occurs in these compartments. These findings have important implications for nutrient sensing and growth control via mTor pathways in metazoans.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
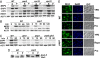
Class C VPS genes are required for efficient induction of Tor-regulated NCR response. (A) Exponentially growing cultures of WT (BY4742) and isogenic class C vps mutants were treated with drug vehicle (−) or 100 nM rapamycin (R) for 30 min or shifted from YPD to proline medium for 1 h (P). RNA was prepared and analyzed by Northern blotting with 32P-labeled DNA probes for GAP1, MEP2, CIT2, SOD2, and ACT1 as loading control. (B) Cultures of WT and pep3 strains grown as in A were shifted into proline medium. Culture aliquots were removed before shifting into proline or after 15, 30, 60, and 120 min, and RNA was isolated and analyzed by Northern blotting with CIT2 and MEP2 probes. (C) Cultures of isogenic WT (BY4742), pep3 (14105), gln3 (10173), and pep3 gln3 (RPY77) strains were shifted from YPD to proline medium for 15 and 60 min. RNA was isolated and GAP1 expression analyzed. (D) Exponentially growing WT (TB123) and pep3 (RPY30) strains were treated as described in A. Gln3-Myc electrophoretic mobility was assayed by Western blot analysis with Myc antibody. (E) Isogenic WT (TB123) and pep3 (RPY30) strains were grown in YPD and treated as in A. Cells were processed for indirect immunofluorescence with anti-Myc. Nuclei were stained with DAPI.
Fig. 2.
Class D vps mutations impair Gln3 function. (A) Isogenic WT, pep3, and the class D vps mutants (pep12, vps45, vps3, vps9, vps19, and vps34) were tested for NCR gene response as in Fig. 1_A_. (B) Isogenic WT and the pep3 and vps45 mutants were assayed for GAP1 expression over 2-h shift from YPD into proline medium. (C) Northern blot signals for GAP1 were quantified in WT (dots), pep3 (squares), and vps45 (triangles) strains and normalized to ACT1 loading control. Results shown are the relative percentage of gene expression with maximal level of expression at 15 min as 100%. (D) Gln3 was detected in WT (TB123) and vps45 (RPY46) strains as in Fig. 1_E_. (E) Percentage of total cells in which Gln3 was nuclear localized in the conditions analyzed in D. Values represent mean ± SD of three independent determinations.
Fig. 3.
A fraction of Gln3 is peripherally associated with light membranes along with Golgi and endosomal markers. (A) Cell-free lysates from isogenic WT (TB123), pep3 (RPY30), and vps45 (RPY46) strains were subjected to differential centrifugation to yield low-speed pellet (P13), supernatant (S13), high-speed pellet (P100), and soluble (S100) fractions. Equal cell equivalents were examined by Western blot to detect Gln3, Vps10, Pep12, and PGK. (B) The WT (TB123), ure2 (RPY64), and the TB123 strain transformed with pURE2 (pVTG20) were fractionated and analyzed by Western blot to detect Ure2 and the Golgi and endosomal markers Kex2 and Pep12 as indicated in Fig. 3_A_. *, Ure2-unrelated polypeptide. (C) S13 fractions from WT (TB123), pep3 (RPY30), and TB123 strain transformed with pURE2 (pVTG20) were incubated with 0.1% Triton X-100, 0.1 M Na2CO3 (pH 11), or 1 M NaCl for 30 min at 4°C. P100 and S100 fractions were isolated and analyzed for Gln3 and Ure2 by Western blot. (D) Schematic representation of the Gln3-Myc9 fusion constructs used in E. The relevant functional domains of Gln3 are shown in the full-length construct 1. (E) The gln3 (10173) mutant was transformed with the Gln3-Myc9 fusion constructs depicted in D. Gln3 in total-cell lysates and P13 and P100 were detected by Western blot. *, Gln3-Myc9 mutant proteins at the predicted molecular masses. Numbers above the fractions correspond to the Gln3-Myc9 construct number in D. Numbers at the right indicate molecular mass in KDa.
Fig. 4.
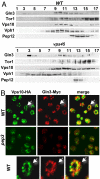
Gln3 cofractionates with Vps10. (A) P100 membranes from WT and vps45 strains were loaded on 18–54% sucrose density step gradients. Gradients were centrifuged and fractionated from top to bottom, and Gln3, Tor1, Vps10, Vph1, and Pep12 were detected by Western blot analysis in equal aliquots from each fraction. (B) Gln3 partially colocalizes with Vps10-containing foci. Cells of WT (RPY73) and pep3 (RPY75), expressing the Gln3-Myc and Vps10-HA fusion proteins from their chromosomal loci, were double stained with monoclonal anti-HA and polyclonal anti-Myc antibodies. Primary antibodies were detected and images were obtained as indicated in Materials and Methods. Arrows in Top indicate a cell magnified in Bottom.
Comment in
- A VAST staging area for regulatory proteins.
Mitchell AP. Mitchell AP. Proc Natl Acad Sci U S A. 2008 May 20;105(20):7111-2. doi: 10.1073/pnas.0803384105. Epub 2008 May 12. Proc Natl Acad Sci U S A. 2008. PMID: 18474868 Free PMC article. No abstract available.
References
- Magasanik B, Kaiser CA. Nitrogen regulation in Saccharomyces cerevisiae. Gene. 2002;290:1–18. - PubMed
- Beck T, Hall MN. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature. 1999;402:689–692. - PubMed
- Bertram PG, et al. Tripartite regulation of Gln3p by TOR, Ure2p, and phosphatases. J Biol Chem. 2000;275:35727–35733. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous