Altered gene expression in the superior temporal gyrus in schizophrenia - PubMed (original) (raw)
Altered gene expression in the superior temporal gyrus in schizophrenia
Nikola A Bowden et al. BMC Genomics. 2008.
Abstract
Background: The superior temporal gyrus (STG), which encompasses the primary auditory cortex, is believed to be a major anatomical substrate for speech, language and communication. The STG connects to the limbic system (hippocampus and amygdala), the thalamus and neocortical association areas in the prefrontal cortex, all of which have been implicated in schizophrenia.
Results: To identify altered mRNA expression in the superior temporal gyrus (STG) in schizophrenia, oligonucleotide microarrays were used with RNA from postmortem STG tissue from 7 individuals with schizophrenia and 7 matched non-psychiatric controls. Overall, there was a trend towards down-regulation in gene expression, and altered expression of genes involved in neurotransmission, neurodevelopment, and presynaptic function was identified. To confirm altered expression identified by microarray analysis, the mRNA expression levels of four genes, IPLA2gamma, PIK31R1, Lin-7b and ATBF1, were semi-quantitatively measured using relative real-time PCR. A number of genes with altered expression in the STG were also shown to have similar changes in expression as shown in our previous study of peripheral blood lymphocytes in schizophrenia.
Conclusion: This study has identified altered expression of genes in the STG involved in neurotransmission and neurodevelopment, and to a lesser extent presynaptic function, which further support the notion of these functions playing an integral role in the development of schizophrenia.
Figures
Figure 1
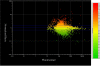
Average ratio of expression of 19,000 genes in the STG from participants with schizophrenia compared to non-psychiatric controls. The central blue line indicates an expression ratio of 1 (ie: equal expression) and the two outer blue lines indicate a 1.5-fold change. Down-regulation is represented as green, up-regulation as red and normal expression as yellow. 191 genes were up-regulated by more than 1.5 fold and 428 were down-regulated by greater than 1.5 fold. The x-axis shows the average level of fluorescence present on the microarrays for each gene.
Figure 2
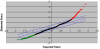
SAM plot of gene expression in the STG from participants with schizophrenia compared to non-psychiatric controls. Average expression of 8737 genes expressed in STG tissue from 7 matched pairs of individuals with schizophrenia and non-psychiatric controls. The central blue line indicates equal expression and the two outer blue lines indicate significantly altered expression (Δ = 0.49, FDR = 0.47), genes in red were significantly up-regulated in schizophrenia compared to controls and genes in green were significantly down-regulated in schizophrenia compared to controls. 216 genes were significantly down-regulated and 85 genes were significantly up-regulated.
Similar articles
- Altered expression of regulator of G-protein signalling 4 (RGS4) mRNA in the superior temporal gyrus in schizophrenia.
Bowden NA, Scott RJ, Tooney PA. Bowden NA, et al. Schizophr Res. 2007 Jan;89(1-3):165-8. doi: 10.1016/j.schres.2006.09.003. Epub 2006 Oct 30. Schizophr Res. 2007. PMID: 17071056 - Microarray analysis of postmortem temporal cortex from patients with schizophrenia.
Aston C, Jiang L, Sokolov BP. Aston C, et al. J Neurosci Res. 2004 Sep 15;77(6):858-66. doi: 10.1002/jnr.20208. J Neurosci Res. 2004. PMID: 15334603 - Altered DARPP-32 expression in the superior temporal gyrus in schizophrenia.
Kunii Y, Yabe H, Wada A, Yang Q, Nishiura K, Niwa S. Kunii Y, et al. Prog Neuropsychopharmacol Biol Psychiatry. 2011 Jun 1;35(4):1139-43. doi: 10.1016/j.pnpbp.2011.03.016. Epub 2011 Mar 29. Prog Neuropsychopharmacol Biol Psychiatry. 2011. PMID: 21453742 - Superior temporal gyrus in schizophrenia: a volumetric magnetic resonance imaging study.
Rajarethinam RP, DeQuardo JR, Nalepa R, Tandon R. Rajarethinam RP, et al. Schizophr Res. 2000 Jan 21;41(2):303-12. doi: 10.1016/s0920-9964(99)00083-3. Schizophr Res. 2000. PMID: 10708339 Review. - Superior temporal gyrus and planum temporale in schizophrenia: a selective review.
Pearlson GD. Pearlson GD. Prog Neuropsychopharmacol Biol Psychiatry. 1997 Nov;21(8):1203-29. doi: 10.1016/s0278-5846(97)00159-0. Prog Neuropsychopharmacol Biol Psychiatry. 1997. PMID: 9460087 Review.
Cited by
- Phosphoinositide-3-kinase regulatory subunit 1 gene polymorphisms are associated with schizophrenia and bipolar disorder in the Han Chinese population.
Huang J, Chen Z, Zhu L, Wu X, Guo X, Yang J, Long J, Su L. Huang J, et al. Metab Brain Dis. 2020 Jun;35(5):785-792. doi: 10.1007/s11011-020-00552-z. Epub 2020 Mar 19. Metab Brain Dis. 2020. PMID: 32193760 - Transcriptome sequencing revealed significant alteration of cortical promoter usage and splicing in schizophrenia.
Wu JQ, Wang X, Beveridge NJ, Tooney PA, Scott RJ, Carr VJ, Cairns MJ. Wu JQ, et al. PLoS One. 2012;7(4):e36351. doi: 10.1371/journal.pone.0036351. Epub 2012 Apr 27. PLoS One. 2012. PMID: 22558445 Free PMC article. - MicroRNA-382 expression is elevated in the olfactory neuroepithelium of schizophrenia patients.
Mor E, Kano S, Colantuoni C, Sawa A, Navon R, Shomron N. Mor E, et al. Neurobiol Dis. 2013 Jul;55:1-10. doi: 10.1016/j.nbd.2013.03.011. Epub 2013 Mar 29. Neurobiol Dis. 2013. PMID: 23542694 Free PMC article. - Medication for Acromegaly Reduces Expression of MUC16, MACC1 and GRHL2 in Pituitary Neuroendocrine Tumour Tissue.
Saksis R, Silamikelis I, Laksa P, Megnis K, Peculis R, Mandrika I, Rogoza O, Petrovska R, Balcere I, Konrade I, Steina L, Stukens J, Breiksa A, Nazarovs J, Sokolovska J, Pirags V, Klovins J, Rovite V. Saksis R, et al. Front Oncol. 2021 Feb 15;10:593760. doi: 10.3389/fonc.2020.593760. eCollection 2020. Front Oncol. 2021. PMID: 33680922 Free PMC article. - Effects of haloperidol and clozapine on synapse-related gene expression in specific brain regions of male rats.
von Wilmsdorff M, Manthey F, Bouvier ML, Staehlin O, Falkai P, Meisenzahl-Lechner E, Schmitt A, Gebicke-Haerter PJ. von Wilmsdorff M, et al. Eur Arch Psychiatry Clin Neurosci. 2018 Sep;268(6):555-563. doi: 10.1007/s00406-018-0872-8. Epub 2018 Feb 5. Eur Arch Psychiatry Clin Neurosci. 2018. PMID: 29404686
References
- Mimmack ML, Ryan M, Baba H, Navarro-Ruiz J, Iritani S, Faull RL, McKenna PJ, Jones PB, Arai H, Starkey M, Emson PC, Bahn S. Gene expression analysis in schizophrenia: reproducible up-regulation of several members of the apolipoprotein L family located in a high-susceptibility locus for schizophrenia on chromosome 22. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:4680–4685. doi: 10.1073/pnas.032069099. - DOI - PMC - PubMed
- Vawter MP, Shannon Weickert C, Ferran E, Matsumoto M, Overman K, Hyde TM, Weinberger DR, Bunney WE, Kleinman JE. Gene expression of metabolic enzymes and a protease inhibitior in the prefrontal cortex are decreased in schizophrenia. Neurochemical Research. 2004;29:1245–1255. doi: 10.1023/B:NERE.0000023611.99452.47. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical