Single bead affinity detection (SINBAD) for the analysis of protein-protein interactions - PubMed (original) (raw)
Single bead affinity detection (SINBAD) for the analysis of protein-protein interactions
Roberta Schulte et al. PLoS One. 2008.
Abstract
We present a miniaturized pull-down method for the detection of protein-protein interactions using standard affinity chromatography reagents. Binding events between different proteins, which are color-coded with quantum dots (QDs), are visualized on single affinity chromatography beads by fluorescence microscopy. The use of QDs for single molecule detection allows the simultaneous analysis of multiple protein-protein binding events and reduces the amount of time and material needed to perform a pull-down experiment.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
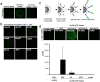
Figure 1. Monitoring protein interactions on a single affinity chromatography bead.
(A) 1 µM recombinant his-TAP-tagged Importin β(wt) was immobilized on 5 ul magnetic Ni-NTA beads for 20 min. After the addition of 5 nM CdSe-ZnS core shell QD655 beads were washed and isolated with a magnet. Beads were recovered in 2 ul PBS, mounted on a glass slide and the bead surface was imaged by confocal microscopy. (B) Schematic illustration of the RanGTP (gray) dissociation of Importin β (red) from Importin α (light blue). (c) QD585 cross-linked to NLS peptide (blue), QD705 cross-linked to reverse NLS (SLN) (magenta), streptavidin-coated QD655-TAP-Importin β(wt) (red) and streptavidin-coated QD605-TAP-Importin β(ΔN44) (green) were incubated with his-Importin α coated Ni-NTA beads in the presence of buffer (control), 20 mM NLS or SLN peptides or 10 mM RanQ69L.
Figure 2. Using SINBAD as pull-down method with increased sensitivity.
(A) Approx. 10 Ni-NTA beads were incubated with reticulocyte lysates containing his-Importin α or his- Importin α together with in vitro translated TAP-Importin β(wt) in the presence or absence of excess Importin α. QD655 were added, beads were isolated and imaged. (B) his-Nup160 was translated in reticulocyte lysates and incubated with equal volume (2 ul) of in vitro translated Nup133-TAP, Nup107-TAP, Nup96-TAP, Nup85-TAP, Nup43-TAP, Nup37-TAP, Seh1-TAP, Sec13-TAP and ELYS-TAP for 30 min. Approx. 5 Ni-NTA beads were added together with streptavidin-coated QDs655 and imaged by confocal microscopy on the bead surface. (C) Schematic illustration of SINBAD for the pull-down of endogenous proteins. (D) Hypotonic cell lysates corresponding to 500 or 50 cells were incubated with Ni-NTA beads coated with his-RanQ69L. Bound endogenous Importin β was visualized as described in (C). (E) Random 10 µm2 areas were imaged and fluorescence intensity at 655 nm was plotted. n = 20.
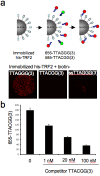
Figure 3. SINBAD for the detection of protein-DNA interactions.
(A) his-TRF2 was immobilized on Ni-NTA beads and incubated with hybridized 5′-biotinylated DNA oligos containing three repeats of TTAGGG (red), a mutant TTACGG or ssTTAGG as indicated. Bound DNA was visualized with streptavidin-coated QDs655. (B) <10 his-TRF2 Ni-NTA beads bound to wild-type telomere sequence were incubated with increasing concentrations of competitor DNA oligos. Fluorescence intensity of QD655 was determined from confocal images and plotted.
Similar articles
- SAINT: probabilistic scoring of affinity purification-mass spectrometry data.
Choi H, Larsen B, Lin ZY, Breitkreutz A, Mellacheruvu D, Fermin D, Qin ZS, Tyers M, Gingras AC, Nesvizhskii AI. Choi H, et al. Nat Methods. 2011 Jan;8(1):70-3. doi: 10.1038/nmeth.1541. Epub 2010 Dec 5. Nat Methods. 2011. PMID: 21131968 Free PMC article. - Affinity-purification coupled to mass spectrometry: basic principles and strategies.
Dunham WH, Mullin M, Gingras AC. Dunham WH, et al. Proteomics. 2012 May;12(10):1576-90. doi: 10.1002/pmic.201100523. Proteomics. 2012. PMID: 22611051 Review. - Isolation of labile multi-protein complexes by in vivo controlled cellular cross-linking and immuno-magnetic affinity chromatography.
Zlatic SA, Ryder PV, Salazar G, Faundez V. Zlatic SA, et al. J Vis Exp. 2010 Mar 9;(37):1855. doi: 10.3791/1855. J Vis Exp. 2010. PMID: 20216526 Free PMC article. - Protein-Protein Interactions: Pull-Down Assays.
Louche A, Salcedo SP, Bigot S. Louche A, et al. Methods Mol Biol. 2017;1615:247-255. doi: 10.1007/978-1-4939-7033-9_20. Methods Mol Biol. 2017. PMID: 28667618 - Resolving protein interactions and complexes by affinity purification followed by label-based quantitative mass spectrometry.
Trinkle-Mulcahy L. Trinkle-Mulcahy L. Proteomics. 2012 May;12(10):1623-38. doi: 10.1002/pmic.201100438. Proteomics. 2012. PMID: 22610586 Review.
Cited by
- Cell cycle-dependent differences in nuclear pore complex assembly in metazoa.
Doucet CM, Talamas JA, Hetzer MW. Doucet CM, et al. Cell. 2010 Jun 11;141(6):1030-41. doi: 10.1016/j.cell.2010.04.036. Cell. 2010. PMID: 20550937 Free PMC article. - Deafness mutation D572N of TMC1 destabilizes TMC1 expression by disrupting LHFPL5 binding.
Yu X, Zhao Q, Li X, Chen Y, Tian Y, Liu S, Xiong W, Huang P. Yu X, et al. Proc Natl Acad Sci U S A. 2020 Nov 24;117(47):29894-29903. doi: 10.1073/pnas.2011147117. Epub 2020 Nov 9. Proc Natl Acad Sci U S A. 2020. PMID: 33168709 Free PMC article. - Probing cellular protein complexes using single-molecule pull-down.
Jain A, Liu R, Ramani B, Arauz E, Ishitsuka Y, Ragunathan K, Park J, Chen J, Xiang YK, Ha T. Jain A, et al. Nature. 2011 May 26;473(7348):484-8. doi: 10.1038/nature10016. Nature. 2011. PMID: 21614075 Free PMC article. - Current Experimental Methods for Characterizing Protein-Protein Interactions.
Zhou M, Li Q, Wang R. Zhou M, et al. ChemMedChem. 2016 Apr 19;11(8):738-56. doi: 10.1002/cmdc.201500495. Epub 2016 Feb 11. ChemMedChem. 2016. PMID: 26864455 Free PMC article. Review. - A Novel Confocal Scanning Protein-Protein Interaction Assay (PPI-CONA) Reveals Exceptional Selectivity and Specificity of CC0651, a Small Molecule Binding Enhancer of the Weak Interaction between the E2 Ubiquitin-Conjugating Enzyme CDC34A and Ubiquitin.
Koszela J, Pham NT, Shave S, St-Cyr D, Ceccarelli DF, Orlicky S, Marinier A, Sicheri F, Tyers M, Auer M. Koszela J, et al. Bioconjug Chem. 2024 Sep 18;35(9):1441-1449. doi: 10.1021/acs.bioconjchem.4c00345. Epub 2024 Aug 21. Bioconjug Chem. 2024. PMID: 39167708 Free PMC article.
References
- Burbulis I, Yamaguchi K, Yu R, Resnekov O, Brent R. Quantifying small numbers of antibodies with a ‘near-universal’ protein-DNA chimera. Nat Methods. 2007;4:1011–1013. - PubMed
- Oh BK, Nam JM, Lee SW, Mirkin CA. A fluorophore-based bio-barcode amplification assay for proteins. Small. 2006;2:103–108. - PubMed
- Patolsky F, Zheng G, Lieber CM. Nanowire-based biosensors. Anal Chem. 2006;78:4260–4269. - PubMed
- Gorlich D, Kutay U. Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Biol. 1999;15:607–660. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources