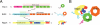Interactions among Ytm1, Erb1, and Nop7 required for assembly of the Nop7-subcomplex in yeast preribosomes - PubMed (original) (raw)
Interactions among Ytm1, Erb1, and Nop7 required for assembly of the Nop7-subcomplex in yeast preribosomes
Lan Tang et al. Mol Biol Cell. 2008 Jul.
Abstract
In Saccharomyces cerevisiae, more than 180 assembly factors associate with preribosomes to enable folding of pre-rRNA, recruitment of ribosomal proteins, and processing of pre-rRNAs to produce mature ribosomes. To examine the molecular architecture of preribosomes and to connect this structure to functions of each assembly factor, assembly subcomplexes have been purified from preribosomal particles. The Nop7-subcomplex contains three assembly factors: Nop7, Erb1, and Ytm1, each of which is necessary for conversion of 27SA(3) pre-rRNA to 27SB(S) pre-rRNA. However, interactions among these three proteins and mechanisms of their recruitment and function in pre-rRNPs are poorly understood. Here we show that Ytm1, Erb1, and Nop7 assemble into preribosomes in an interdependent manner. We identified which domains within Ytm1, Erb1, and Nop7 are necessary for their interaction with each other and are sufficient for recruitment of each protein into preribosomes. Dominant negative effects on growth and ribosome biogenesis caused by overexpressing truncated Ytm1, Erb1, or Nop7 constructs, and recessive phenotypes of the truncated proteins revealed not only interaction domains but also other domains potentially important for each protein to function in ribosome biogenesis. Our data suggest a model for the architecture of the Nop7-subcomplex and provide potential functions of domains of each protein.
Figures
Figure 1.
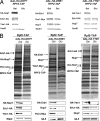
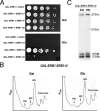
Interdependence of assembly of Erb1, Nop7, and Ytm1 into preribosomes. Yeast strains containing GAL-NOP7, GAL-ERB1, or GAL-YTM1 were grown at 30°C in galactose medium to 3·107 cells/ml. A second culture of each strain was grown in galactose medium and shifted to glucose medium for 16 h to 3 × 107 cells/ml, to deplete the respective proteins. Preribosomes were isolated from whole cell extracts by affinity purification, using Rpf2-TAP. (A) Proteins in whole cell extracts were assayed by Western blotting. (B) Proteins in affinity-purified preribosomes were resolved by SDS-PAGE and stained with silver (top) or subjected to Western blot analysis (bottom). Positions of stained Erb1, Nop7, and Ytm1 are indicated.
Figure 2.
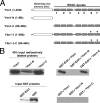
The C-terminal WD-40 domain of Ytm1 (i.e., Ytm1-C) interacts directly with Erb1. (A) Schematic representation of Ytm1 and truncation constructs. Shown are the notchless-like element (NLE, white rectangle), and WD40 repeats 1–7 (hatched boxes). Black triangles (▾) indicate two mutations: G398D and S442N. (B) Left, 40% of the input [35S] labeled proteins in each assay and 100% of the pulled down GST proteins (Coomassie blue staining) are shown. Right, synthetic 35S-methionine–labeled full-length Ytm1, Ytm1-N, or Ytm1-C proteins (*) were incubated with purified GST-Erb1 protein. Complexes were eluted from glutathione beads, subjected to SDS-PAGE, and detected by autoradiography.
Figure 3.
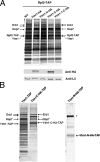
Ytm1-C assembles into preribosomes and the Nop7-subcomplex in vivo. Yeast strains containing each of the _GAL_-inducible full-length or truncated YTM1 genes were grown at 30°C in synthetic medium containing 1% raffinose to ∼1.5·107 cells/ml. Galactose was added to each strain to a final concentration of 1% to induce the expression of Ytm1 proteins. Cells were harvested after 4-h induction (one doubling time). (A) Lysates were prepared from cells expressing endogenous Ytm1 plus HA-tagged full-length Ytm1, Ytm1-N, Ytm1-C, or Ytm1-1-C, or from cells expressing only endogenous Ytm1 (WT). Pre-rRNPs were purified using TAP-tagged Rpf2. Proteins present in the affinity-purified preribosomes were resolved by SDS-PAGE and stained with silver (top) or subjected to Western blot analysis (bottom) using anti-HA or anti-rpL5 antiserum. (B) Left, preribosomes purified from wild-type cells using TAP-tagged Ytm1 and purified from cells using TAP-tagged Ytm1-C. Right, preribosomes purified from cells expressing TAP-tagged Ytm1-N. Proteins in affinity-purified samples were resolved by SDS-PAGE and stained with silver. Protein bands in the Ytm1-C-TAP purified samples were excised and subjected to mass spectrometry to identify Erb1, Nop7, and Ytm1-C-TAP. The band indicated by an asterisk is Ste23, a protein not necessary for ribosome biogenesis (L. Tang, unpublished data).
Figure 4.
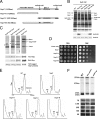
Expression of Ytm1-C causes dominant negative effects on growth and ribosome biogenesis. (A) Wild-type cells or yeast expressing endogenous Ytm1 plus full-length or truncated Ytm1 proteins were grown to early log phase, serially diluted (10-, 100-, 1000 and 10,000-fold) onto Glu (glucose) or Gal (galactose) medium, and incubated at 30°C. (B) Wild-type cells or yeast overexpressing Ytm1-C or co-overexpressing Ytm1-C and full-length Ytm1 were grown to early log phase and serially diluted as above. (C) Whole cell extracts were subjected to centrifugation on sucrose gradients to separate 40S and 60S ribosomal subunits, 80S ribosomes, and polyribosomes. (D) Pre-rRNA processing pathway in the yeast S. cerevisiae. (E) RNA was extracted from wild-type yeast (WT) or from strains in which Ytm1 was depleted (GAL-YTM1) or Ytm1-C was overexpressed (YTM1-C). Pre-rRNAs were assayed by primer extension (left) or by Northern blotting (right), using specific oligonucleotide primers or probes, respectively.
Figure 5.
The N-terminal conserved region of Erb1 interacts with both Nop7 and Ytm1 and assembles into preribosomes. (A) Schematic representation of full-length and truncated Erb1 constructs. Shown are the N-terminal conserved region of Erb1 (white rectangle) and the WD40 repeats 1–7 (hatched boxes). (B) Lysates were prepared from cells expressing endogenous Erb1 plus HA-tagged full-length Erb1 or each Erb1 truncation or from wild-type cells only expressing endogenous Erb1 (WT), as in Figure 3. Pre-rRNPs were purified using TAP-tagged Rpf2. Proteins present in the affinity-purified preribosomes were resolved by SDS-PAGE and stained with silver (top) or subjected to Western blot analysis (bottom) using antibodies against the HA tag, or ribosomal protein L5. (C) TAP-tagged Erb1-N, Erb1-C1, Erb1-C2, or Erb1-M was expressed using the GAL promoter by the addition of galactose to a final concentration of 1% for 4 h. Proteins associated with each tagged, truncated protein were affinity-purified, resolved by SDS-PAGE, and stained with silver (top) or subjected to Western blot analysis using antibodies against Nop7, the Myc tag on Ytm1, or the HA tag on the Erb1 proteins (bottom). The black arrow (→) in each panel indicates the corresponding TAP-tagged Erb1 truncation.
Figure 6.
Expression of Erb1-C2 causes dominant negative effects on growth and ribosome biogenesis. (A) Wild-type cells or yeast expressing endogenous Erb1 plus full-length or truncated Erb1 proteins were grown to early log phase and serially diluted as in Figure 4A. (B) Extracts from wild-type cells (WT) or from cells overexpressing the indicated forms of Erb1 were subjected to centrifugation on sucrose gradients to separate 40S and 60S ribosomal subunits, 80S ribosomes, and polyribosomes. (C) RNA was extracted from wild-type yeast (WT) or from strains in which Erb1 was depleted (GAL-ERB1) or Erb1-C2 was overexpressed (ERB1-C2). Pre-rRNAs were assayed by primer extension (left) or Northern blotting (right), using specific oligonucleotide primers or probes, respectively.
Figure 7.
The C-terminal WD40 domain of Erb1 is not essential for growth or ribosome assembly. (A) Empty vector or a plasmid containing each ERB1 truncation driven by the ERB1 promoter was introduced into the GAL-ERB1 strain. Cells were grown to early log phase in galactose medium, serially diluted (10-, 100-, 1000-, and 10,000-fold) onto Glu (glucose) medium, to turn off the wild-type GAL-ERB1 gene, or Gal (galactose) medium, to express GAL-ERB1, and incubated at 30°C. (B) Erb1-N was expressed from its own promoter in a GAL-ERB1 strain. This GAL-ERB1 ERB1-N strain was grown at 30°C in galactose medium to 3 ×107 cells/ml. A second culture was grown in galactose medium and shifted to glucose medium for 16 h to 3 ×107 cells/ml, to deplete wild-type Erb1 protein. Whole cell extracts were subjected to centrifugation on sucrose gradients to separate 40S and 60S ribosomal subunits, 80S ribosomes, and polyribosomes. (C) RNA was extracted from the GAL-ERB1 ERB1-N strain grown in galactose, or shifted to glucose for 16 h. The 27SA2, 27SA3, 27SBL, and 27SBS pre-rRNAs present in each strain were identified by primer extension.
Figure 8.
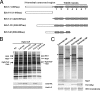
Interaction domains of Nop7. (A) Schematic representation of Nop7 and its truncation constructs. Shown are the N-terminal pescadillo domain of Nop7 (white rectangle), BRCT domain (dotted box), and two coil-coiled domains (gray boxes). (B) Pre-rRNPs were purified from cells expressing endogenous Nop7 plus HA-tagged full-length Nop7 or each Nop7 truncation or from wild-type cells only expressing endogenous Nop7 (WT) as in Figure 3. Proteins present in the affinity-purified preribosomes were resolved by SDS-PAGE and stained with silver (top), or subjected to Western blot analysis (bottom) using antibodies against the HA epitope, or ribosomal protein rpL5. (C) TAP-tagged Nop7-N, Nop7-C, Nop7-M, or full-length Nop7 was expressed from the GAL promoter by the addition of galactose to a final concentration of 1% for 4 h. Cell extracts from these strains were prepared and subjected to a high-speed spin (42K) for 3 h. Proteins associated with each tagged truncation were affinity-purified from the supernatants, resolved by SDS-PAGE, and stained with silver (top), or subjected to Western blot analysis using antibodies against the Myc tag on Ytm1 or Erb1, or the protein A tag on Nop7-TAP (bottom). The black arrow in each panel indicates the corresponding TAP-tagged Nop7 truncation. The asterisk (*) indicates Ste23, a protein not necessary for ribosome biogenesis (L. Tang, unpublished data). (D) Wild-type cells or yeast expressing full-length or truncated Nop7 proteins were grown to early log phase and serially diluted as in Figure 4A. (E) Cell extracts were subjected to centrifugation on sucrose gradients to separate 40S and 60S ribosomal subunits, 80S ribosomes, and polyribosomes. (F) RNA was extracted from wild-type yeast (WT) or from strains in which Nop7 was depleted (GAL-NOP7) or Nop7-M was overexpressed (NOP7-M). Pre-rRNAs were assayed by primer extension (top) or Northern blotting (bottom), using specific oligonucleotide primers or probes, respectively.
Figure 9.
(A) Interaction domains within the Nop7-subcomplex. Light pink or light green boxes in each protein indicate the smallest region shown to interact with their full-length ligands. (B) Architecture of the Nop7-subcomplex.
Similar articles
- Ytm1, Nop7, and Erb1 form a complex necessary for maturation of yeast 66S preribosomes.
Miles TD, Jakovljevic J, Horsey EW, Harnpicharnchai P, Tang L, Woolford JL Jr. Miles TD, et al. Mol Cell Biol. 2005 Dec;25(23):10419-32. doi: 10.1128/MCB.25.23.10419-10432.2005. Mol Cell Biol. 2005. PMID: 16287855 Free PMC article. - Concerted removal of the Erb1-Ytm1 complex in ribosome biogenesis relies on an elaborate interface.
Thoms M, Ahmed YL, Maddi K, Hurt E, Sinning I. Thoms M, et al. Nucleic Acids Res. 2016 Jan 29;44(2):926-39. doi: 10.1093/nar/gkv1365. Epub 2015 Dec 10. Nucleic Acids Res. 2016. PMID: 26657628 Free PMC article. - The Crystal Structure of the Ubiquitin-like Domain of Ribosome Assembly Factor Ytm1 and Characterization of Its Interaction with the AAA-ATPase Midasin.
Romes EM, Sobhany M, Stanley RE. Romes EM, et al. J Biol Chem. 2016 Jan 8;291(2):882-93. doi: 10.1074/jbc.M115.693259. Epub 2015 Nov 24. J Biol Chem. 2016. PMID: 26601951 Free PMC article. - Assembly factors Rpf2 and Rrs1 recruit 5S rRNA and ribosomal proteins rpL5 and rpL11 into nascent ribosomes.
Zhang J, Harnpicharnchai P, Jakovljevic J, Tang L, Guo Y, Oeffinger M, Rout MP, Hiley SL, Hughes T, Woolford JL Jr. Zhang J, et al. Genes Dev. 2007 Oct 15;21(20):2580-92. doi: 10.1101/gad.1569307. Genes Dev. 2007. PMID: 17938242 Free PMC article. - Eukaryotic Ribosome Assembly.
Baßler J, Hurt E. Baßler J, et al. Annu Rev Biochem. 2019 Jun 20;88:281-306. doi: 10.1146/annurev-biochem-013118-110817. Epub 2018 Dec 19. Annu Rev Biochem. 2019. PMID: 30566372 Review.
Cited by
- Ribosomal proteins L7 and L8 function in concert with six A₃ assembly factors to propagate assembly of domains I and II of 25S rRNA in yeast 60S ribosomal subunits.
Jakovljevic J, Ohmayer U, Gamalinda M, Talkish J, Alexander L, Linnemann J, Milkereit P, Woolford JL Jr. Jakovljevic J, et al. RNA. 2012 Oct;18(10):1805-22. doi: 10.1261/rna.032540.112. Epub 2012 Aug 14. RNA. 2012. PMID: 22893726 Free PMC article. - The N-terminal extension of yeast ribosomal protein L8 is involved in two major remodeling events during late nuclear stages of 60S ribosomal subunit assembly.
Tutuncuoglu B, Jakovljevic J, Wu S, Gao N, Woolford JL Jr. Tutuncuoglu B, et al. RNA. 2016 Sep;22(9):1386-99. doi: 10.1261/rna.055798.115. Epub 2016 Jul 7. RNA. 2016. PMID: 27390266 Free PMC article. - Ribosome biogenesis in the yeast Saccharomyces cerevisiae.
Woolford JL Jr, Baserga SJ. Woolford JL Jr, et al. Genetics. 2013 Nov;195(3):643-81. doi: 10.1534/genetics.113.153197. Genetics. 2013. PMID: 24190922 Free PMC article. Review. - The AAA-ATPase Rea1 drives removal of biogenesis factors during multiple stages of 60S ribosome assembly.
Bassler J, Kallas M, Pertschy B, Ulbrich C, Thoms M, Hurt E. Bassler J, et al. Mol Cell. 2010 Jun 11;38(5):712-21. doi: 10.1016/j.molcel.2010.05.024. Mol Cell. 2010. PMID: 20542003 Free PMC article. - A cluster of ribosome synthesis factors regulate pre-rRNA folding and 5.8S rRNA maturation by the Rat1 exonuclease.
Granneman S, Petfalski E, Tollervey D. Granneman S, et al. EMBO J. 2011 Aug 2;30(19):4006-19. doi: 10.1038/emboj.2011.256. EMBO J. 2011. PMID: 21811236 Free PMC article.
References
- Alber F., et al. Determining the architectures of macromolecular assemblies. Nature. 2007a;450:683–694. - PubMed
- Alber F., et al. The molecular architecture of the nuclear pore complex. Nature. 2007b;450:695–701. - PubMed
- Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G., Smith J. A., Struhl K. Current Protocols in Molecular Biology. New York: John Wiley & Sons; 1994.
- Boisvert F. M., van Koningsbruggen S., Navascues J., Lamond A. I. The multifunctional nucleolus. Nat. Rev. Mol. Cell Biol. 2007;8:574–585. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases