3.88 A structure of cytoplasmic polyhedrosis virus by cryo-electron microscopy - PubMed (original) (raw)
. 2008 May 15;453(7193):415-9.
doi: 10.1038/nature06893. Epub 2008 Apr 30.
Affiliations
- PMID: 18449192
- PMCID: PMC2746981
- DOI: 10.1038/nature06893
3.88 A structure of cytoplasmic polyhedrosis virus by cryo-electron microscopy
Xuekui Yu et al. Nature. 2008.
Abstract
Cytoplasmic polyhedrosis virus (CPV) is unique within the Reoviridae family in having a turreted single-layer capsid contained within polyhedrin inclusion bodies, yet being fully capable of cell entry and endogenous RNA transcription. Biochemical data have shown that the amino-terminal 79 residues of the CPV turret protein (TP) is sufficient to bring CPV or engineered proteins into the polyhedrin matrix for micro-encapsulation. Here we report the three-dimensional structure of CPV at 3.88 A resolution using single-particle cryo-electron microscopy. Our map clearly shows the turns and deep grooves of alpha-helices, the strand separation in beta-sheets, and densities for loops and many bulky side chains; thus permitting atomic model-building effort from cryo-electron microscopy maps. We observed a helix-to-beta-hairpin conformational change between the two conformational states of the capsid shell protein in the region directly interacting with genomic RNA. We have also discovered a messenger RNA release hole coupled with the mRNA capping machinery unique to CPV. Furthermore, we have identified the polyhedrin-binding domain, a structure that has potential in nanobiotechnology applications.
Figures
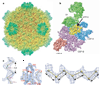
Figure 1. Overall structure of the CPV capsid
a, Radially coloured, shaded surface view of the CPV reconstruction as viewed along a two-fold axis. b, An extracted asymmetric unit, colour coded by protein subunits, including one turret protein (TP) (green), two copies of the capsid shell proteins (CSP-A in blue and CSP-B in purple), and two copies of the large protrusion protein (LPP-5, near the five-fold axis, in yellow; LPP-3, near the three-fold axis, in brown). c, d, Views of density maps of one α-helix (c) and four β-strands (d) for CSP-B superimposed with the corresponding Cα model, showing the clear turn and deep groove of the α-helix with a pitch of ~5.8Å and the density for the bulky side chain of residue 520 in c, and the clear separation of β-strands in d (note also the densities for the bulky side chains of amino acids 341 and 343). e, Cα model of a β-strand of TP in the density map, showing zigzagging of the backbone and the clear densities for side chains. All the inter-consecutive Cα distances are ~3.8Å , which is a well established distance in polypeptide chains.
Figure 2. A conformational change between CSP-A and CSP-B: implication for packing and sliding of the dsRNA genome
a, Atomic model of CSP-A, coloured from blue at the N terminus to red at the C terminus. b, Close-up view of the CSP-A (blue) and CSP-B (purple) density map, showing that an α-helix (the upper helix within the green density) in CSP-A transforms into part of a β-hairpin in CSP-B (the upper part in the yellow density). c, Density maps with Cα models, showing the conformational change between CSP-A (left) and CSP-B (right). One α-helix in CSP-A transforms into part of the β-hairpin in CSP-B (indicated by curved arrow). The changing path is also indicated by an empty arrow. d, A 10Å slab extracted from the two-fold map showing the ordered dsRNA genome with a ~27Å distance between the adjacent dsRNA strands (arrow). The dotted box indicates the A-spike plug all the way in the central chamber of the turret.
Figure 3. Nascent mRNA release hole coupled with the GTase active site of TP in a way unique to CPV
a, Close-up view of the atomic model of CSP-A (two copies) and CSP-B. The mRNA release hole comprises loops contributed by the two copies of CSP-A (red and orange) and one CSP-B (cyan). b, c, Cα models of one CSP-B (cyan), two copies of CSP-A (red and orange) and two GTase domains (blue and light blue), showing the mRNA releasing and capping pathway, which is illustrated by green arrows in b (viewed from outside of the capsid) and c (viewed from inside of the capsid). d, Cα models of the GTase domains in a turret, showing that the active-site cleft (yellow stars) of GTase rotates away from the central chamber, facing sideways, thus coupled with the mRNA release hole (white circle).
Figure 4. Location and structure of the unique polyhedrin-binding domain (PBD) of TP
a, Shaded surface representation of TP viewed from inside, showing the two methylase domains (purple), GTase domain (blue), and CPV’s unique PBD (orange). The PBD is absent from other dsRNA viruses and consists of residues 1–79 of TP. b, Ribbon model of the CPV polyhedrin trimer, which is the building block of the crystalline polyhedra. c, View of one entire turret, showing the orientation and position of one TP (coloured) in the turret. d, Stereo view of a cryo-electron microscopy density map superimposed with the Cα model of GTase domain (green) and PBD (blue), which are connected together through a linker. e, Blown-up view of the model of PBD superimposed with its cryo-electron microscopy density.
Similar articles
- Atomic model of a cypovirus built from cryo-EM structure provides insight into the mechanism of mRNA capping.
Cheng L, Sun J, Zhang K, Mou Z, Huang X, Ji G, Sun F, Zhang J, Zhu P. Cheng L, et al. Proc Natl Acad Sci U S A. 2011 Jan 25;108(4):1373-8. doi: 10.1073/pnas.1014995108. Epub 2011 Jan 10. Proc Natl Acad Sci U S A. 2011. PMID: 21220303 Free PMC article. - Atomic model of CPV reveals the mechanism used by this single-shelled virus to economically carry out functions conserved in multishelled reoviruses.
Yu X, Ge P, Jiang J, Atanasov I, Zhou ZH. Yu X, et al. Structure. 2011 May 11;19(5):652-61. doi: 10.1016/j.str.2011.03.003. Structure. 2011. PMID: 21565700 Free PMC article. - In situ structures of the segmented genome and RNA polymerase complex inside a dsRNA virus.
Zhang X, Ding K, Yu X, Chang W, Sun J, Zhou ZH. Zhang X, et al. Nature. 2015 Nov 26;527(7579):531-534. doi: 10.1038/nature15767. Epub 2015 Oct 26. Nature. 2015. PMID: 26503045 Free PMC article. - Towards atomic resolution structural determination by single-particle cryo-electron microscopy.
Zhou ZH. Zhou ZH. Curr Opin Struct Biol. 2008 Apr;18(2):218-28. doi: 10.1016/j.sbi.2008.03.004. Epub 2008 Apr 9. Curr Opin Struct Biol. 2008. PMID: 18403197 Free PMC article. Review. - Captivating Perplexities of Spinareovirinae 5' RNA Caps.
Kniert J, Lin QF, Shmulevitz M. Kniert J, et al. Viruses. 2021 Feb 13;13(2):294. doi: 10.3390/v13020294. Viruses. 2021. PMID: 33668598 Free PMC article. Review.
Cited by
- Predictive modeling and cryo-EM: A synergistic approach to modeling macromolecular structure.
Corum MR, Venkannagari H, Hryc CF, Baker ML. Corum MR, et al. Biophys J. 2024 Feb 20;123(4):435-450. doi: 10.1016/j.bpj.2024.01.021. Epub 2024 Jan 23. Biophys J. 2024. PMID: 38268190 Review. - Cryo-electron tomography to study viral infection.
Graham M, Zhang P. Graham M, et al. Biochem Soc Trans. 2023 Aug 31;51(4):1701-1711. doi: 10.1042/BST20230103. Biochem Soc Trans. 2023. PMID: 37560901 Free PMC article. Review. - Interaction between Bombyx mori Cytoplasmic Polyhedrosis Virus NSP8 and BmAgo2 Inhibits RNA Interference and Enhances Virus Proliferation.
Pan J, Qiu Q, Kumar D, Xu J, Tong X, Shen Z, Zhu M, Hu X, Gong C. Pan J, et al. Microbiol Spectr. 2023 Aug 17;11(4):e0493822. doi: 10.1128/spectrum.04938-22. Epub 2023 Jun 21. Microbiol Spectr. 2023. PMID: 37341621 Free PMC article. - Cryo-EM structure-based selection of computed ligand poses enables design of MTA-synergic PRMT5 inhibitors of better potency.
Zhou W, Yadav GP, Yang X, Qin F, Li C, Jiang QX. Zhou W, et al. Commun Biol. 2022 Oct 3;5(1):1054. doi: 10.1038/s42003-022-03991-9. Commun Biol. 2022. PMID: 36192627 Free PMC article. - 3D electron diffraction for structure determination of small-molecule nanocrystals: A possible breakthrough for the pharmaceutical industry.
Andrusenko I, Gemmi M. Andrusenko I, et al. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2022 Sep;14(5):e1810. doi: 10.1002/wnan.1810. Epub 2022 May 20. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2022. PMID: 35595285 Free PMC article. Review.
References
- Mertens PPC, Attoui H, Duncan R, Dermody TS. In: Virus Taxonomy: Eighth Report of the International Committee on Taxonomy of Viruses. Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA, editors. London: Elsevier/Academic Press; 2005. pp. 447–454.
- Zhou ZH. In: Segmented Double-Stranded RNA Viruses: Structure and Molecular Biology. Patton JT, editor. Norfolk: Caister Academic Press; 2008. pp. 27–43.
- Hill CL, et al. The structure of a cypovirus and the functional organization of dsRNA viruses. Nature Struct. Biol. 1999;6:565–568. - PubMed
- Ikeda K, et al. Immobilization of diverse foreign proteins in viral polyhedra and potential application for protein microarrays. Proteomics. 2006;6:54–66. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM071940-01A2/GM/NIGMS NIH HHS/United States
- R01 GM071940-04/GM/NIGMS NIH HHS/United States
- R01 AI069015-01A1/AI/NIAID NIH HHS/United States
- P41 RR002250-217385/RR/NCRR NIH HHS/United States
- P41 RR002250-200043/RR/NCRR NIH HHS/United States
- R01 GM071940-02/GM/NIGMS NIH HHS/United States
- P41 RR002250/RR/NCRR NIH HHS/United States
- P41 RR002250-190043/RR/NCRR NIH HHS/United States
- R01 AI069015-02/AI/NIAID NIH HHS/United States
- P41 RR002250-226489/RR/NCRR NIH HHS/United States
- R01 AI069015/AI/NIAID NIH HHS/United States
- R01 GM071940-03/GM/NIGMS NIH HHS/United States
- R01 AI069015-03/AI/NIAID NIH HHS/United States
- R01 GM071940/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases