Pygopus activates Wingless target gene transcription through the mediator complex subunits Med12 and Med13 - PubMed (original) (raw)
Pygopus activates Wingless target gene transcription through the mediator complex subunits Med12 and Med13
Inés Carrera et al. Proc Natl Acad Sci U S A. 2008.
Abstract
Wnt target gene transcription is mediated by nuclear translocation of stabilized beta-catenin, which binds to TCF and recruits Pygopus, a cofactor with an unknown mechanism of action. The mediator complex is essential for the transcription of RNA polymerase II-dependent genes; it associates with an accessory subcomplex consisting of the Med12, Med13, Cdk8, and Cyclin C subunits. We show here that the Med12 and Med13 subunits of the Drosophila mediator complex, encoded by kohtalo and skuld, are essential for the transcription of Wingless target genes. kohtalo and skuld act downstream of beta-catenin stabilization both in vivo and in cell culture. They are required for transcriptional activation by the N-terminal domain of Pygopus, and their physical interaction with Pygopus depends on this domain. We propose that Pygopus promotes Wnt target gene transcription by recruiting the mediator complex through interactions with Med12 and Med13.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
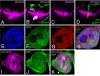
skd and kto are required for the expression of Wg target genes. Third instar wing discs (A–H and K–P) and third instar eye disc (I and J) are shown. Anterior is to the left and dorsal is up in this and subsequent figures. The phenotypes of skd and kto mutations are indistinguishable (30, 31), so only one genotype is shown in each experiment. Clones homozygous for skdT413 (A–F), ktoT241 (G and H), skdT606 (I and J), ktoT555 (K and L), cdk8K185 (M and N), or cycCY5 (O and P) are marked by the absence of GFP (green in B, D, F, H, J, L, N, and P). (A and B) Wg staining in magenta; the arrow in B indicates a clone at the wing margin. (C, D, O, and P) β-gal staining reflects Dll-lacZ expression in magenta. (E, F, M, and N) Sens staining is in magenta. (G and H) β-gal staining reflects vgQ-lacZ expression in magenta. (I and J) β-gal staining reflects ds-lacZ expression in magenta. (K and L) β-gal staining reflects sal-lacZ expression in magenta. Although Wg is still expressed at the wing margin in skd or kto mutant clones, expression of Wg target genes is lost. However, Wg target genes are unaffected in cdk8 or cycC mutant clones.
Fig. 2.
skd and kto act downstream of Arm and Pygo. Third instar wing discs (A–D) and third instar antennal discs (E–K) are shown. (A and B) Clones expressing ArmΔN are marked by coexpression of GFP (green in B). (C and D) Clones expressing ArmΔN and homozygous for ktoT631 are marked by coexpression of GFP (green in D). β-gal staining reflecting Dll-lacZ expression is shown in magenta in A–D. ArmΔN can activate Dll expression in wild-type, but not kto mutant, cells. (E–H) HA staining (blue in E and H) shows expression of _hs_-HAPygoGAL4 after a 2-h heat shock and a 2-h recovery. UAS-GFP expression is shown in green in F and H. Clones homozygous for skdT413 are marked by a lack of arm-lacZ expression (β-gal staining in red in G and H). Loss of skd does not affect the expression of _hs_-HAPygoGAL4, but abolishes its ability to activate UAS-GFP. (I and K) Clones homozygous for ktoT555 (arrowheads) are marked by lack of arm-lacZ expression (β-gal staining in magenta in I and K). UAS-GFP expression driven by _tub_-GAL4 is shown in green in J and K. GAL4 can activate UAS-GFP in the absence of kto.
Fig. 3.
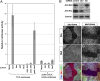
skd and kto are required for the expression of a Wg reporter in cultured cells. (A) Ratio of TCF firefly luciferase to the transfection control Pol III-RL in Kc cells treated with the indicated dsRNAs. The TCF luciferase reporter is strongly activated when axin is knocked down by RNAi, but this activation is reduced 80- to 100-fold by knocking down skd or kto in addition to axin. In Kc cells transfected with _actin_-GAL4 and UAS-luciferase, knocking down skd or kto reduces activation of the reporter by ≈3-fold. Error bars indicate the standard deviation between the triplicate samples tested for each dsRNA. This figure is a representative example of three independent experiments. (B) Western blot showing the levels of Skd and Kto protein in Kc cells treated with cdk8 (control), skd, or kto dsRNA. skd knockdown also partially reduces the level of Kto protein. The bottom blot shows a band that cross-reacts with the Kto antibody and serves as a loading control. (C–H) Wing imaginal discs with clones homozygous for ktoT241 (C–E) or skdT606 (F–H) marked by the absence of GFP (green in E and H) and stained with anti-Kto (C and F; blue in E and H) and anti-Skd (D and G; red in E and H). Kto protein is reduced in skd mutant cells, but Skd protein is unaffected in kto mutant cells.
Fig. 4.
Pygo physically interacts with Skd. (A) Anti-Skd immunoprecipitations (IPs) of extracts from embryos expressing UAS-HAPygo or UAS-HAPygoΔNHD (58) with the ubiquitous drivers daughterless (da)-GAL4 or tubulin (tub)-GAL4. Input lanes show 1% of the input for the IP. HAPygoΔNHD is less efficiently coimmunoprecipitated with anti-Skd than full-length Pygo. The lower blot shows that the unrelated nuclear protein PCNA does not coimmunoprecipitate with anti-Skd. (B) Coimmunoprecipitation of HAPygo with anti-Skd from Kc cells treated with lacZ or skd dsRNA. Removing Skd protein greatly reduces Pygo coimmunoprecipitation, demonstrating the specificity of the Skd antibody. Input lanes show 0.5% of the input, and control lanes show IPs with Protein A beads but no primary antibody. (C) Coimmunoprecipitation of a Pygo construct that lacks the PHD domain, HA-PygoΔPHDGAL4, with anti-Skd. Input lane shows 1% of the input, and the control lane shows an IP with no primary antibody. This figure shows that the interaction with Skd is independent of Pygo binding to the Lgs/Arm/TCF complex. (D) Model consistent with our results. Pygo, one of the most downstream components of the Wg-responsive transcriptional complex, may recruit the mediator complex through interactions of its NHD with Skd/Med13 and Kto/Med12, leading to transcriptional activation of Wg target genes. The C-terminal domain of Arm also directly interacts with Med12 (63), enhancing binding to the mediator complex; this interaction may explain why skd and kto have a stronger effect than pygo on Wg target genes.
Similar articles
- A complex of Armadillo, Legless, and Pygopus coactivates dTCF to activate wingless target genes.
Thompson BJ. Thompson BJ. Curr Biol. 2004 Mar 23;14(6):458-66. doi: 10.1016/j.cub.2004.02.026. Curr Biol. 2004. PMID: 15043810 - A role of Pygopus as an anti-repressor in facilitating Wnt-dependent transcription.
Mieszczanek J, de la Roche M, Bienz M. Mieszczanek J, et al. Proc Natl Acad Sci U S A. 2008 Dec 9;105(49):19324-9. doi: 10.1073/pnas.0806098105. Epub 2008 Nov 26. Proc Natl Acad Sci U S A. 2008. PMID: 19036929 Free PMC article. - Dissecting nuclear Wingless signalling: recruitment of the transcriptional co-activator Pygopus by a chain of adaptor proteins.
Städeli R, Basler K. Städeli R, et al. Mech Dev. 2005 Nov;122(11):1171-82. doi: 10.1016/j.mod.2005.07.004. Epub 2005 Aug 22. Mech Dev. 2005. PMID: 16169192 - Pygopus and the Wnt signaling pathway: a diverse set of connections.
Jessen S, Gu B, Dai X. Jessen S, et al. Bioessays. 2008 May;30(5):448-56. doi: 10.1002/bies.20757. Bioessays. 2008. PMID: 18404694 Review. - The Yin-Yang of TCF/beta-catenin signaling.
Barker N, Morin PJ, Clevers H. Barker N, et al. Adv Cancer Res. 2000;77:1-24. doi: 10.1016/s0065-230x(08)60783-6. Adv Cancer Res. 2000. PMID: 10549354 Review.
Cited by
- Genetic Screen in Adult Drosophila Reveals That dCBP Depletion in Glial Cells Mitigates Huntington Disease Pathology through a Foxo-Dependent Pathway.
Martin E, Heidari R, Monnier V, Tricoire H. Martin E, et al. Int J Mol Sci. 2021 Apr 9;22(8):3884. doi: 10.3390/ijms22083884. Int J Mol Sci. 2021. PMID: 33918672 Free PMC article. - The functions of Mediator in Candida albicans support a role in shaping species-specific gene expression.
Uwamahoro N, Qu Y, Jelicic B, Lo TL, Beaurepaire C, Bantun F, Quenault T, Boag PR, Ramm G, Callaghan J, Beilharz TH, Nantel A, Peleg AY, Traven A. Uwamahoro N, et al. PLoS Genet. 2012;8(4):e1002613. doi: 10.1371/journal.pgen.1002613. Epub 2012 Apr 5. PLoS Genet. 2012. PMID: 22496666 Free PMC article. - Defining the essential function of FBP/KSRP proteins: Drosophila Psi interacts with the mediator complex to modulate MYC transcription and tissue growth.
Guo L, Zaysteva O, Nie Z, Mitchell NC, Amanda Lee JE, Ware T, Parsons L, Luwor R, Poortinga G, Hannan RD, Levens DL, Quinn LM. Guo L, et al. Nucleic Acids Res. 2016 Sep 19;44(16):7646-58. doi: 10.1093/nar/gkw461. Epub 2016 May 20. Nucleic Acids Res. 2016. PMID: 27207882 Free PMC article. - Wnt/beta-catenin signaling: components, mechanisms, and diseases.
MacDonald BT, Tamai K, He X. MacDonald BT, et al. Dev Cell. 2009 Jul;17(1):9-26. doi: 10.1016/j.devcel.2009.06.016. Dev Cell. 2009. PMID: 19619488 Free PMC article. Review. - Beta-catenin hits chromatin: regulation of Wnt target gene activation.
Mosimann C, Hausmann G, Basler K. Mosimann C, et al. Nat Rev Mol Cell Biol. 2009 Apr;10(4):276-86. doi: 10.1038/nrm2654. Nat Rev Mol Cell Biol. 2009. PMID: 19305417 Review.
References
- Bjorklund S, Gustafsson CM. The yeast Mediator complex and its regulation. Trends Biochem Sci. 2005;30:240–244. - PubMed
- Conaway RC, Sato S, Tomomori-Sato C, Yao T, Conaway JW. The mammalian Mediator complex and its role in transcriptional regulation. Trends Biochem Sci. 2005;30:250–255. - PubMed
- Kim YJ, Lis JT. Interactions between subunits of Drosophila Mediator and activator proteins. Trends Biochem Sci. 2005;30:245–249. - PubMed
- Thompson CM, Koleske AJ, Chao DM, Young RA. A multisubunit complex associated with the RNA polymerase II CTD and TATA-binding protein in yeast. Cell. 1993;73:1361–1375. - PubMed
- Kim YJ, Bjorklund S, Li Y, Sayre MH, Kornberg RD. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell. 1994;77:599–608. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases