Characterization of two functional NKX3.1 binding sites upstream of the PCAN1 gene that are involved in the positive regulation of PCAN1 gene transcription - PubMed (original) (raw)
Characterization of two functional NKX3.1 binding sites upstream of the PCAN1 gene that are involved in the positive regulation of PCAN1 gene transcription
Wenwen Liu et al. BMC Mol Biol. 2008.
Abstract
Background: NKX3.1 and PCAN1 are both prostate-specific genes related to prostate development and prostate cancer. So far, little is known about the regulatory mechanisms of the expression of these two genes. In the present study, we found that NKX3.1 upregulated PCAN1 gene transcription in LNCaP prostate cancer cells. To understand the regulatory mechanisms, our work focused on identifying the functional NKX3.1 binding sites upstream of the PCAN1 gene, which might be involved in the positive regulation of PCAN1 expression by NKX3.1.
Results: We cloned and characterized a 2.6 kb fragment upstream of the PCAN1 gene. Analysis of the 2.6 kb sequence with MatInspector 2.2 revealed five potential binding sites of NKX3.1 transcription factor. Luciferase reporter assays, electrophoretic mobility shift assays, chromatin immunoprecipitation and RNA interference were performed to study the effects of NKX3.1 on PCAN1 gene expression in prostate cancer cells. Our results showed that PCAN1 promoter activity and mRNA expression were increased by transfection with the NKX3.1 containing plasmid (pcDNA3.1-NKX3.1) and that PCAN1 mRNA expression was decreased by RNA interference targeting human NKX3.1 in LNCaP prostate cancer cells. The results of electrophoretic mobility shift assays and chromatin immunoprecipitation showed that NKX3.1 bound to NBS1 (-1848 to -1836) and NBS3 (-803 to -791) upstream of the PCAN1 gene. The luciferase reporter assays showed that NBS1 and NBS3 enhanced the promoter activity in pGL3-promoter vector with cotransfection of the NKX3.1 containing plasmid. Furthermore, the deletion of NBS1 or both NBS1 and NBS3 reduced PCAN1 promoter activity and abolished the positive regulation of PCAN1 expression by NKX3.1.
Conclusion: Our results suggested that two functional NKX3.1 binding sites located at -1848 to -1836 and -803 to -791 upstream of the PCAN1 gene were involved in the positive regulation of PCAN1 gene transcription by NKX3.1.
Figures
Figure 1
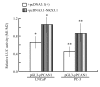
Effects of NKX3.1 on the PCAN1 promoter in LNCaP and PC-3 cells. LNCaP and PC-3 cells were cotransfected with pGL3-_p_PCAN1 and pcDNA3.1-NKX3.1 (grey bars). The control cells were cotransfected with pGL3-_p_PCAN1 and pcDNA3.1 (+) plasmid (blank bars). The cells were harvested 48 h after transfection and PCAN1 promoter activity was detected by dual-luciferase reporter assays. Results were expressed as relative luciferase activities (M1/M2). The data were represented as the mean of four individual values ± SD.*p < 0.05, **p < 0.05, grey bar vs blank bar.
Figure 2
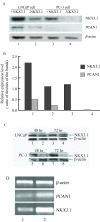
Effects of NKX3.1 on PCAN1 mRNA expression in LNCaP and PC-3 cells. The effects of NKX3.1 on PCAN1 mRNA expression as detected by RT-PCR. A. PCAN1 mRNA expression following transfection of LNCaP cells with 1. pcDNA3.1-NKX3.1; 2. pcDNA3.1 (+); or following transfection of PC-3 cells with 3. pcDNA3.1-NKX3.1; or 4. pcDNA3.1 (+). B. Relative expression levels were presented as the ratio of densities of NKX3.1 or PCAN1 to β-actin bands. The results were expressed as mean ± SD (n = 3). C. The expression of NKX3.1 protein in LNCaP and PC-3 cells after transfection of pcDNA3.1-NKX3.1 (1, 3, 5, 7) or pcDNA3.1 (+) (2, 4, 6, 8).D. NKX3.1 and PCAN1 mRNA expression following stable transfection of LNCaP cells with 1. pRNAT-RNAi1 targeting human NKX3.1; 2. pRNAT-RiN as a control vector.
Figure 3
Binding assays of NKX3.1 binding sites with nuclear extracts. A. Locations and sequences of five potential NKX3.1 binding sites (NBSs) upstream of the PCAN1 gene. B. EMSA was performed to assay the binding activities of the five NBSs with the nuclear extracts from LNCaP cells. 1–5: labelled NBS1~NBS5 with LNCaP nuclear extracts respectively. A DNA-protein binding complex was formed when the labelled probe of NBS1 (lane 1) or NBS3 (lane 3) was reacted with LNCaP nuclear extracts. C and D. Competition binding and antibody blocking were used to detect the specific binding of the NBS1 (C) or NBS3 (D) with NKX3.1. Labelled NBS1 or NBS3 without nuclear extracts was used as the control (lane1); a specific DNA-protein complex was formed when labelled NBS1 or NBS3 was reacted with LNCaP nuclear extracts (lane 2). Competition binding was performed in the presence of a 250-fold molar excess of unlabelled NBS1 or NBS3 (lane 3), unlabelled mutant NBS1 or NBS3 (lane 4). The binding activity of probe NBS1 or NBS3 to nuclear extracts could be blocked by anti-NKX3.1 antibody (lane5).
Figure 4
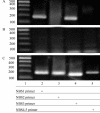
NKX3.1 binds to the NBSs of the PCAN1 promoter in living cells. LNCaP cells transfected with pcDNA3.1-NKX3.1 were cross-linked by formaldehyde treatment and lysed. Cell lysates were subjected to immunoprecipitation with either an antibody to NKX3.1 (A) or rabbit IgG (B). Four primers (names and sequences are shown in Table 1) spanning five NBSs of PCAN1 promoter region were used for PCR of recovered DNA from the immunoprecipitation (lanes 2–5). Input DNA was used as positive control (C).
Figure 5
Effects of NBSs on the SV40 heterogeneous promoter. The sequences of five NBSs were synthesized in vitro and inserted upstream of the SV40 promoter in pGL3-promoter vector generating the pGL3-NBS-promoter plasmids. The five recombinant pGL3-NBS-promoter plasmids or pGL3-promoter vector were then cotransfected with pcDNA3.1-NKX3.1 respectively, to study the effects of NBSs on the activity of the SV40 heterogeneous promoter. Cotransfection of pGL3-NBS-promoter with pcDNA3.1 (+) vector was used as the control. The promoter activities were determined by dual luciferase assays. Results were expressed as relative luciferase activity (M1/M2). The data were represented as the mean of four individual values ± SD. *p < 0.01,**p < 0.01, strip bar vs blank bar.
Figure 6
Effects of deletion of NBS1 and NBS3 on PCAN1 promoter activity. pGL3-_p_PCAN1 is a luciferase reporter plasmid containing a 2.6 kb wild type PCAN1 promoter; pGL3-NBS-Id represented pGL3-NBS1id-_p_PCAN1 with deletion of NBS1 from the PCAN1 promoter; pGL3-NBS-Idd represented pGL3-NBS1,3idd-_p_PCAN1, in which both NBS1 and NBS3 were deleted from the PCAN1 promoter. They were cotransfected with pcDNA3.1-NKX3.1 (grey bar) or pcDNA3.1 (+) vector (blank bar) into LNCaP cells, and the promoter activity was analyzed by dual-luciferase reporter assays. Results were expressed as relative luciferase activities (M1/M2). The data were represented as the mean of four individual values ± SD. *p < 0.01, **p < 0.05, grey bar vs blank bar.
Similar articles
- Molecular cloning and analysis of the human PCAN1 (GDEP) promoter.
Liu W, Chen W, Zhang P, Yu C, Kong F, Deng J, Zhang J, Jiang A. Liu W, et al. Cell Mol Biol Lett. 2007;12(4):482-92. doi: 10.2478/s11658-007-0016-z. Epub 2007 Apr 28. Cell Mol Biol Lett. 2007. PMID: 17468839 Free PMC article. - Identification of an inhibitory cis-element in the promoter region of human homeobox gene NKX3.1.
Anli J, Pengju Z, Xiaoyan H, Wenwei C, Zhifang L, Feng K, Huiqing Y, Jianye Z. Anli J, et al. Prostate. 2006 Sep 1;66(12):1339-46. doi: 10.1002/pros.20330. Prostate. 2006. PMID: 16845664 - Identification of Sp1-elements in the promoter region of human homeobox gene NKX3.1.
Yu CX, Jin T, Chen WW, Zhang PJ, Liu WW, Guan HY, Zhang J, Liu QW, Jiang AL. Yu CX, et al. Mol Biol Rep. 2009 Nov;36(8):2353-60. doi: 10.1007/s11033-009-9457-y. Epub 2009 Mar 5. Mol Biol Rep. 2009. PMID: 19263243 - Loss of the NKX3.1 tumorsuppressor promotes the TMPRSS2-ERG fusion gene expression in prostate cancer.
Thangapazham R, Saenz F, Katta S, Mohamed AA, Tan SH, Petrovics G, Srivastava S, Dobi A. Thangapazham R, et al. BMC Cancer. 2014 Jan 13;14:16. doi: 10.1186/1471-2407-14-16. BMC Cancer. 2014. PMID: 24418414 Free PMC article. - Androgen regulation of the prostatic tumour suppressor NKX3.1 is mediated by its 3' untranslated region.
Thomas MA, Preece DM, Bentel JM. Thomas MA, et al. Biochem J. 2010 Jan 15;425(3):575-83. doi: 10.1042/BJ20091109. Biochem J. 2010. PMID: 19886863
Cited by
- Gene expression profiles in the PC-3 human prostate cancer cells induced by NKX3.1.
Zhang P, Liu W, Zhang J, Guan H, Chen W, Cui X, Liu Q, Jiang A. Zhang P, et al. Mol Biol Rep. 2010 Mar;37(3):1505-12. doi: 10.1007/s11033-009-9549-8. Epub 2009 May 22. Mol Biol Rep. 2010. PMID: 19462257 - IGFBP3, a transcriptional target of homeobox D10, is correlated with the prognosis of gastric cancer.
Xue M, Fang Y, Sun G, Zhuo W, Zhong J, Qian C, Wang L, Wang L, Si J, Chen S. Xue M, et al. PLoS One. 2013 Dec 27;8(12):e81423. doi: 10.1371/journal.pone.0081423. eCollection 2013. PLoS One. 2013. PMID: 24386080 Free PMC article. - Transcription factor CCAAT/enhancer binding protein alpha up-regulates microRNA let-7a-1 in lung cancer cells by direct binding.
Lin Y, Zhao J, Hu X, Wang L, Liang L, Chen W. Lin Y, et al. Cancer Cell Int. 2016 Mar 9;16:17. doi: 10.1186/s12935-016-0294-5. eCollection 2016. Cancer Cell Int. 2016. PMID: 26962302 Free PMC article. - Nucleosome dynamics define transcriptional enhancers.
He HH, Meyer CA, Shin H, Bailey ST, Wei G, Wang Q, Zhang Y, Xu K, Ni M, Lupien M, Mieczkowski P, Lieb JD, Zhao K, Brown M, Liu XS. He HH, et al. Nat Genet. 2010 Apr;42(4):343-7. doi: 10.1038/ng.545. Epub 2010 Mar 7. Nat Genet. 2010. PMID: 20208536 Free PMC article.
References
- Jemal A, Siegel R, Ward E, Murray T, Xu J, Smigal C, Thun MJ. Cancer statistics, 2006. CA Cancer J Clin. 2006;56:106–130. - PubMed
- Saad F, Al Dejmah A, Perrotte P, McCormack M, Benard F, Valiquette L, Karakiewicz PI. Therapeutic approach to hormone-refractory prostate cancer. Can J Urol. 2006;13:52–56. - PubMed
- Di Lorenzo G, De Placido S. Hormone refractory prostate cancer (HRPC): present and future approaches of therapy. Int J Immunopathol Pharmacol. 2006;19:11–34. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous