Malignant transformation initiated by Mll-AF9: gene dosage and critical target cells - PubMed (original) (raw)
Malignant transformation initiated by Mll-AF9: gene dosage and critical target cells
Weili Chen et al. Cancer Cell. 2008 May.
Abstract
The pathways by which oncogenes, such as MLL-AF9, initiate transformation and leukemia in humans and mice are incompletely defined. In a study of target cells and oncogene dosage, we found that Mll-AF9, when under endogenous regulatory control, efficiently transformed LSK (Lin(-)Sca1(+)c-kit(+)) stem cells, while committed granulocyte-monocyte progenitors (GMPs) were transformation resistant and did not cause leukemia. Mll-AF9 was expressed at higher levels in hematopoietic stem (HSC) than GMP cells. Mll-AF9 gene dosage effects were directly shown in experiments where GMPs were efficiently transformed by the high dosage of Mll-AF9 resulting from retroviral transduction. Mll-AF9 upregulated expression of 192 genes in both LSK and progenitor cells, but to higher levels in LSKs than in committed myeloid progenitors.
Figures
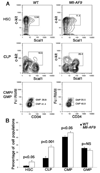
Figure 1. Mll-AF9 results in the expansion of HSC, CLP and CMP populations
a, Sorting profiles of HSCs, CLPs, CMPs and GMPs showing the expansion of c-kit+Sca1+ HSCs (in Lin−IL-7R− population) and CLP (in Lin−IL-7R+ population) in Mll-AF9 mice. Expansion of the CMP population is also shown. b, Significantly higher percentages of HSCs, CLPs and CMPs were found in lineage negative marrow cells of Mll-AF9 than those of wild type mice. Error bars represent the standard error of the means.
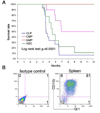
Figure 2. HSCs and CLPs are more efficient than CMPs and GMPs in producing leukemia in vivo
**a,**Survival of irradiated mice receiving 100–2500 cells. The survival rate was calculated using Kaplan-Meier analysis. The HSC and CLP groups had significantly poorer survival than the CMP and GMP groups (p<0.0001, log-rank test). b, Recipient mice developed myeloid leukemia after transplantation. All recipient mice showed the same high CD11b+Gr1+ profile in spleen previously described in Mll-AF9 leukemic mice. FACS on the spleen of a representative recipient is shown.
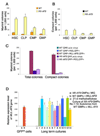
Figure 3. In Mll-AF9 knockin mice, HSCs and CLPs produce more total and compact myeloid colonies with enhanced self-renewal in vitro than CMPs and GMPs. MLL-AF9 retrovirally transduced GMPs produce the most total and compact myeloid colonies
a, Total myeloid colony numbers were higher in HSCs and CLPs than in CMPs and GMPs in Mll-AF9 mice after 21-day culture under myeloid conditions. All the Mll-AF9 populations had significantly increased colonies compared to WT. b. Compact colony numbers were higher in HSCs/CLPs than in CMPs/GMPs in Mll-AF9 mice after 21-day culture. No compact colonies were detected in WT mice. Error bars represent standard error of the means. c, Total myeloid colony and compact colony numbers were the highest in MLL-AF9 retrovirally transduced GMP cells compared to the MIG (MSCV-IRES-GFP) vector transduced knockin Mll-AF9 GMPs and other knockin Mll-AF9 GMP controls as labeled. d, MLL-AF9 expression in retrovirally transduced GMPs and knockin Mll-AF9 GMPs. Error bars represent standard error of the means.
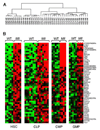
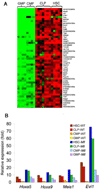
Figure 4. Hierarchical clustering of Mll-AF9 LSK (HSC/CLP), and CMP/GMP populations. Genes overexpressed in HSC, CLP, CMP and GMP populations
a, A two-way ANOVA with stratified permutation testing was performed to select genes differentially expressed in Mll-AF9 compared to wild type cells in each of the four populations. Hierarchical clustering performed with the 446 genes selected by the two-way ANOVA (FDR<0.1) separates the HSCs/CLPs from the CMPs/GMPs. WT = wild type, _Mll_=_Mll-AF9_. **b,** _Mll-AF9_ up-regulates expression of genes in multiple cell types (FDR<0.1, two-way ANOVA with permutation testing). Heat-maps showing the expression level of the top 50 genes up-regulated in the _Mll-AF9_ samples compared to WT, ranked in decreasing order of fold change up-regulation in HSCs. Expression levels are represented by colors: black=median red > median, green < median. Gene identifiers are at right.
Figure 5. A set of genes are up-regulated by Mll-AF9 to highest levels in LSKs (HSCs/CLPs) compared to both to Mll-AF9 CMPs/GMPs and to wild type HSCs/CLPs
a, Mll-AF9 up-regulated genes are highly expressed in HSCs and CLPs compared to CMPs and GMPs. Of the 192 genes over-expressed in Mll-AF9 cells, 96 genes are expressed at higher levels in HSCs and CLPs compared to CMPs and GMPs (FDR<0.1, SAM). The top 50 genes in this subset are shown. Expression levels are represented by colors: black = median, red > median, green < median. b, Expression of Mll-AF9 up-regulated genes – Hoxa5, Hoxa9, Meis1 and Evi1 is highest in Mll-AF9 HSCs/CLPs. Data represent average expression relative to levels in wild type GMPs.
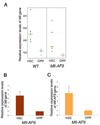
Figure 6. Mll-AF9 expression is higher in HSC than GMP populations. Retrovirally transduced MLL-AF9 results in very high expression levels of the oncogene
a, Mll expression in the HSCs and GMPs from WT and Mll-AF9 mice by microarray. b, Mll expression in the HSCs and GMPs from Mll-AF9 mice by real-time RT-PCR. c, Mll-AF9 expression in the HSCs and GMPs from Mll-AF9 mice by real-time RT-PCR. d, MLL-AF9 expression in retrovirally transduced GMPs and knockin Mll-AF9 GMPs. Error bars represent standard error of the means.
Similar articles
- MLL-AF9 and FLT3 cooperation in acute myelogenous leukemia: development of a model for rapid therapeutic assessment.
Stubbs MC, Kim YM, Krivtsov AV, Wright RD, Feng Z, Agarwal J, Kung AL, Armstrong SA. Stubbs MC, et al. Leukemia. 2008 Jan;22(1):66-77. doi: 10.1038/sj.leu.2404951. Epub 2007 Sep 13. Leukemia. 2008. PMID: 17851551 Free PMC article. - EVI1 is critical for the pathogenesis of a subset of MLL-AF9-rearranged AMLs.
Bindels EM, Havermans M, Lugthart S, Erpelinck C, Wocjtowicz E, Krivtsov AV, Rombouts E, Armstrong SA, Taskesen E, Haanstra JR, Beverloo HB, Döhner H, Hudson WA, Kersey JH, Delwel R, Kumar AR. Bindels EM, et al. Blood. 2012 Jun 14;119(24):5838-49. doi: 10.1182/blood-2011-11-393827. Epub 2012 May 2. Blood. 2012. PMID: 22553314 Free PMC article. - ZNF521 Enhances MLL-AF9-Dependent Hematopoietic Stem Cell Transformation in Acute Myeloid Leukemias by Altering the Gene Expression Landscape.
Chiarella E, Aloisio A, Scicchitano S, Todoerti K, Cosentino EG, Lico D, Neri A, Amodio N, Bond HM, Mesuraca M. Chiarella E, et al. Int J Mol Sci. 2021 Oct 6;22(19):10814. doi: 10.3390/ijms221910814. Int J Mol Sci. 2021. PMID: 34639154 Free PMC article. - Learning from mouse models of MLL fusion gene-driven acute leukemia.
Schwaller J. Schwaller J. Biochim Biophys Acta Gene Regul Mech. 2020 Aug;1863(8):194550. doi: 10.1016/j.bbagrm.2020.194550. Epub 2020 Apr 19. Biochim Biophys Acta Gene Regul Mech. 2020. PMID: 32320749 Review. - MLL translocations, histone modifications and leukaemia stem-cell development.
Krivtsov AV, Armstrong SA. Krivtsov AV, et al. Nat Rev Cancer. 2007 Nov;7(11):823-33. doi: 10.1038/nrc2253. Nat Rev Cancer. 2007. PMID: 17957188 Review.
Cited by
- Meis1 preserves hematopoietic stem cells in mice by limiting oxidative stress.
Unnisa Z, Clark JP, Roychoudhury J, Thomas E, Tessarollo L, Copeland NG, Jenkins NA, Grimes HL, Kumar AR. Unnisa Z, et al. Blood. 2012 Dec 13;120(25):4973-81. doi: 10.1182/blood-2012-06-435800. Epub 2012 Oct 22. Blood. 2012. PMID: 23091297 Free PMC article. - Acute myeloid leukemia stem cells and CD33-targeted immunotherapy.
Walter RB, Appelbaum FR, Estey EH, Bernstein ID. Walter RB, et al. Blood. 2012 Jun 28;119(26):6198-208. doi: 10.1182/blood-2011-11-325050. Epub 2012 Jan 27. Blood. 2012. PMID: 22286199 Free PMC article. Review. - Induction of autophagy and inhibition of melanoma growth in vitro and in vivo by hyperactivation of oncogenic BRAF.
Maddodi N, Huang W, Havighurst T, Kim K, Longley BJ, Setaluri V. Maddodi N, et al. J Invest Dermatol. 2010 Jun;130(6):1657-67. doi: 10.1038/jid.2010.26. Epub 2010 Feb 25. J Invest Dermatol. 2010. PMID: 20182446 Free PMC article. - PIM serine/threonine kinases in the pathogenesis and therapy of hematologic malignancies and solid cancers.
Brault L, Gasser C, Bracher F, Huber K, Knapp S, Schwaller J. Brault L, et al. Haematologica. 2010 Jun;95(6):1004-15. doi: 10.3324/haematol.2009.017079. Epub 2010 Feb 9. Haematologica. 2010. PMID: 20145274 Free PMC article. Review. - AMPK activation induces immunogenic cell death in AML.
Mondesir J, Ghisi M, Poillet L, Bossong RA, Kepp O, Kroemer G, Sarry JE, Tamburini J, Lane AA. Mondesir J, et al. Blood Adv. 2023 Dec 26;7(24):7585-7596. doi: 10.1182/bloodadvances.2022009444. Blood Adv. 2023. PMID: 37903311 Free PMC article.
References
- Akashi K, Traver D, Miyamoto T, Weissman IL. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;6774:193–197. - PubMed
- Barjesteh van Waalwijk van Doorn-Khosrovani S, Erpelinck C, Lowenberg B, Delwel R. Low expression of MDS1-EVI1-like-1 (MEL1) and EVI1-like-1 (EL1) genes in favorable-risk acute myeloid leukemia. Exp. Hematol. 2003;11:1066–1072. - PubMed
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Roy. Statistical Society. 1995:289–300.
- Caslini C, Serna A, Rossi V, Introna M, Biondi A. Modulation of cell cycle by graded expression of MLL-AF4 fusion oncoprotein. Leukemia. 2004;6:1064–1071. - PubMed
- Chapiro E, Russell L, Radford-Weiss I, Bastard C, Lessard M, Struski S, Cave H, Fert-Ferrer S, Barin C, Maarek O, et al. Overexpression of CEBPA resulting from the translocation t(14;19)(q32;q13) of human precursor B acute lymphoblastic leukemia. Blood. 2006;10:3560–3563. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA087053-07/CA/NCI NIH HHS/United States
- R01-CA087053/CA/NCI NIH HHS/United States
- K08 CA122191/CA/NCI NIH HHS/United States
- R01 CA087053-05/CA/NCI NIH HHS/United States
- R01 CA087053-04/CA/NCI NIH HHS/United States
- K08-CA122191/CA/NCI NIH HHS/United States
- R01 CA087053/CA/NCI NIH HHS/United States
- R01 CA087053-06/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases