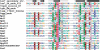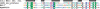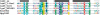Structural and evolutionary classification of Type II restriction enzymes based on theoretical and experimental analyses - PubMed (original) (raw)
Structural and evolutionary classification of Type II restriction enzymes based on theoretical and experimental analyses
Jerzy Orlowski et al. Nucleic Acids Res. 2008 Jun.
Abstract
For a very long time, Type II restriction enzymes (REases) have been a paradigm of ORFans: proteins with no detectable similarity to each other and to any other protein in the database, despite common cellular and biochemical function. Crystallographic analyses published until January 2008 provided high-resolution structures for only 28 of 1637 Type II REase sequences available in the Restriction Enzyme database (REBASE). Among these structures, all but two possess catalytic domains with the common PD-(D/E)XK nuclease fold. Two structures are unrelated to the others: R.BfiI exhibits the phospholipase D (PLD) fold, while R.PabI has a new fold termed 'half-pipe'. Thus far, bioinformatic studies supported by site-directed mutagenesis have extended the number of tentatively assigned REase folds to five (now including also GIY-YIG and HNH folds identified earlier in homing endonucleases) and provided structural predictions for dozens of REase sequences without experimentally solved structures. Here, we present a comprehensive study of all Type II REase sequences available in REBASE together with their homologs detectable in the nonredundant and environmental samples databases at the NCBI. We present the summary and critical evaluation of structural assignments and predictions reported earlier, new classification of all REase sequences into families, domain architecture analysis and new predictions of three-dimensional folds. Among 289 experimentally characterized (not putative) Type II REases, whose apparently full-length sequences are available in REBASE, we assign 199 (69%) to contain the PD-(D/E)XK domain. The HNH domain is the second most common, with 24 (8%) members. When putative REases are taken into account, the fraction of PD-(D/E)XK and HNH folds changes to 48% and 30%, respectively. Fifty-six characterized (and 521 predicted) REases remain unassigned to any of the five REase folds identified so far, and may exhibit new architectures. These enzymes are proposed as the most interesting targets for structure determination by high-resolution experimental methods. Our analysis provides the first comprehensive map of sequence-structure relationships among Type II REases and will help to focus the efforts of structural and functional genomics of this large and biotechnologically important class of enzymes.
Figures
Figure 1.
Clustering of Type II REase sequences and their assignment to three-dimensional folds. (A) Representative structures of nuclease domains of Type II REases or proteins sharing the same fold: PD-(D/E)XK: BamHI (3bam); the universally conserved core is indicated in green, nonconserved structures in gray, HNH: catalytic domain of T4 endonuclease VII (1en7), PLD: catalytic domain of R.BfiI (2c1l), GIY-YIG: catalytic domain of homing endonuclease I-TevI (1mk0), HALFPIPE: R.PabI (2dvy). (B) Results of clustering of Type II REases from REBASE and their homologs in the nr and env_nr database with CLANS (with promiscuous domains, such as MTase or GHKL domains, excluded from analysis). Structures in (A) and sequences in (B) are colored according to the their assignment to fold families (see below): PD-D(E)XK: green, HNH: blue, GIY-YIG: yellow, PLD: magenta, HALFPIPE: cyan, unclassified: red. Connections between dots represent the degree of pairwise sequence similarity, as quantified by BLAST _P_-value (the darker the line, the higher similarity). The whole ‘galaxy’ of REases is held together by a certain level of ‘background’ similarity between different (often unrelated) sequences that is due to pure chance. Thus, while connections within dense clusters practically always reflect high similarity and evolutionary relationship, connections between clusters do not have to reflect their phylogenetic relationships (although they often do, especially in the case of close connections with multiple dark lines). All subfamilies with >20 members or with representatives with solved X-ray structures have been labeled by the name of their representative sequence.
Figure 2.
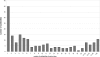
The distribution of size (number of members) among REase subfamilies. Seventy-seven subfamilies (41% of all subfamilies) contain < 5 sequences, which makes it very difficult to analyze the patterns of sequence conservation and e.g. identify invariant residues that could form active sites.
Figure 3.
Sequence alignment of representative Type II REases from all subfamilies of the PD-(D/E)XK superfamily. Sequences of REases are preceded with sequences of several proteins from this superfamily with solved crystal structures and with typical secondary structure representation (of 1gef Holiday junction resolvase). Amino acids are colored according to physico-chemical properties of their side chains (negatively charged: red; positively charged: blue, violet; hydrophilic: gray; hydrophobic: green, magenta, yellow). Residues with more than 50% sequence conservation are shaded. Nonconserved sequence linkers between conserved blocks have been omitted for clarity.
Figure 4.
Sequence alignment of representative Type II REases from all subfamilies of the HNH superfamily. Sequences of REases are preceded with sequences of several proteins from this superfamily with solved crystal structures and with typical secondary structure representation (of 1en7 T4 endonuclease VII). Amino acids are colored according to physico-chemical properties of their side chains (negatively charged: red; positively charged: blue, violet; hydrophilic: gray; hydrophobic: green, magenta, yellow). Residues with more than 50% sequence conservation are shaded.
Figure 5.
Sequence alignment of representative Type II REases from the PLD superfamily. Sequences of REases are preceded with a sequence of Nuc nuclease (1BYR) from the PLD superfamily and with the secondary structure of R.BfiI (2c1l). Amino acids are colored according to physico-chemical properties of their side chains (negatively charged: red; positively charged: blue, violet; hydrophilic: gray; hydrophobic: green, magenta, yellow). Residues with more than 70% sequence conservation are shaded.
Figure 6.
Sequence alignment of representative Type II REases from the GIY-YIG superfamily. Sequences of two REases are preceded by sequences of GIY-YIG members with solved crystal structures and with the secondary structure of I-TevI homing endonuclease (1mk0). Amino acids are colored according to physico-chemical properties of their side chains (negatively charged: red; positively charged: blue, violet; hydrophilic: gray; hydrophobic: green, magenta, yellow). Residues with more than 70% sequence conservation are shaded. Nonconserved sequence linkers between conserved blocks have been omitted for clarity.
Figure 7.
A variety of primary structures (domain architectures on the sequence level) in confirmed and putative Type II REases. Sequences are aligned by their nuclease domains. Drawing in scale, length of PD-D(E)XK domain corresponds to 110 aa. Some very long sequences are broken for the clarity of presentation.
Figure 8.
Fraction of enzymes assigned to different folds, purged at maximum 90% identity. (A) Confirmed REases from REBASE; (B) putative REASES from REBASE; (C) putative REASES from REBASE and all homologs found nonredundant (nr) and environmental samples (env_nr) NCBI database.
Figure 9.
Number of Type II REases from different folds leaving 5′ or 3′ overhangs of different length or blunt ends.
Similar articles
- Type II restriction endonuclease R.Hpy188I belongs to the GIY-YIG nuclease superfamily, but exhibits an unusual active site.
Kaminska KH, Kawai M, Boniecki M, Kobayashi I, Bujnicki JM. Kaminska KH, et al. BMC Struct Biol. 2008 Nov 14;8:48. doi: 10.1186/1472-6807-8-48. BMC Struct Biol. 2008. PMID: 19014591 Free PMC article. - Type II restriction endonuclease R.Eco29kI is a member of the GIY-YIG nuclease superfamily.
Ibryashkina EM, Zakharova MV, Baskunov VB, Bogdanova ES, Nagornykh MO, Den'mukhamedov MM, Melnik BS, Kolinski A, Gront D, Feder M, Solonin AS, Bujnicki JM. Ibryashkina EM, et al. BMC Struct Biol. 2007 Jul 12;7:48. doi: 10.1186/1472-6807-7-48. BMC Struct Biol. 2007. PMID: 17626614 Free PMC article. - Theoretical model of restriction endonuclease HpaI in complex with DNA, predicted by fold recognition and validated by site-directed mutagenesis.
Skowronek KJ, Kosinski J, Bujnicki JM. Skowronek KJ, et al. Proteins. 2006 Jun 1;63(4):1059-68. doi: 10.1002/prot.20920. Proteins. 2006. PMID: 16498623 - Crystallographic and bioinformatic studies on restriction endonucleases: inference of evolutionary relationships in the "midnight zone" of homology.
Bujnicki JM. Bujnicki JM. Curr Protein Pept Sci. 2003 Oct;4(5):327-37. doi: 10.2174/1389203033487072. Curr Protein Pept Sci. 2003. PMID: 14529527 Review. - Categoric prediction of metal ion mechanisms in the active sites of 17 select type II restriction endonucleases.
Advani S, Mishra P, Dubey S, Thakur S. Advani S, et al. Biochem Biophys Res Commun. 2010 Nov 12;402(2):177-9. doi: 10.1016/j.bbrc.2010.09.113. Epub 2010 Oct 1. Biochem Biophys Res Commun. 2010. PMID: 20888795 Review.
Cited by
- Tetrameric structure of the restriction DNA glycosylase R.PabI in complex with nonspecific double-stranded DNA.
Wang D, Miyazono KI, Tanokura M. Wang D, et al. Sci Rep. 2016 Oct 12;6:35197. doi: 10.1038/srep35197. Sci Rep. 2016. PMID: 27731370 Free PMC article. - Nucleases: diversity of structure, function and mechanism.
Yang W. Yang W. Q Rev Biophys. 2011 Feb;44(1):1-93. doi: 10.1017/S0033583510000181. Epub 2010 Sep 21. Q Rev Biophys. 2011. PMID: 20854710 Free PMC article. Review. - Characterization and genomic analysis of a novel Synechococcus phage S-H9-2 belonging to Bristolvirus genus isolated from the Yellow Sea.
Luo L, Ma X, Guo R, Jiang T, Wang T, Shao H, He H, Wang H, Liang Y, McMinn A, Guo C, Wang M. Luo L, et al. Virus Res. 2023 Apr 15;328:199072. doi: 10.1016/j.virusres.2023.199072. Epub 2023 Feb 26. Virus Res. 2023. PMID: 36781075 Free PMC article. - Structure, subunit organization and behavior of the asymmetric Type IIT restriction endonuclease BbvCI.
Shen BW, Doyle L, Bradley P, Heiter DF, Lunnen KD, Wilson GG, Stoddard BL. Shen BW, et al. Nucleic Acids Res. 2019 Jan 10;47(1):450-467. doi: 10.1093/nar/gky1059. Nucleic Acids Res. 2019. PMID: 30395313 Free PMC article. - Live virus-free or die: coupling of antivirus immunity and programmed suicide or dormancy in prokaryotes.
Makarova KS, Anantharaman V, Aravind L, Koonin EV. Makarova KS, et al. Biol Direct. 2012 Nov 14;7:40. doi: 10.1186/1745-6150-7-40. Biol Direct. 2012. PMID: 23151069 Free PMC article.
References
- Skowronek KJ, Bujnicki JM. In: Industrial Enzymes: Structure, Function and Applications. Polaina J, MacCabe AP, editors. Springer; 2007. Chapter 21.
- Williams RJ. Restriction endonucleases: classification, properties, and applications. Mol. Biotechnol. 2003;23:225–243. - PubMed
- Pingoud AM. Restriction Endonucleases. Berlin, Heidelberg: Springer; 2004.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases