Site-specific release of nascent chains from ribosomes at a sense codon - PubMed (original) (raw)
Site-specific release of nascent chains from ribosomes at a sense codon
Victoria A Doronina et al. Mol Cell Biol. 2008 Jul.
Abstract
"2A" oligopeptides are autonomous elements containing a D(V/I)EXNPGP motif at the C terminus. Protein synthesis from an open reading frame containing an internal 2A coding sequence yields two separate polypeptides, corresponding to sequences up to and including 2A and those downstream. We show that the 2A reaction occurs in the ribosomal peptidyltransferase center. Ribosomes pause at the end of the 2A coding sequence, over the glycine and proline codons, and the nascent chain up to and including this glycine is released. Translation-terminating release factors eRF1 and eRF3 play key roles in the reaction. On the depletion of eRF1, a greater proportion of ribosomes extend through the 2A coding sequence, yielding the full-length protein. In contrast, impaired eRF3 GTPase activity leads to many ribosomes failing to translate beyond 2A. Further, high-level expression of a 2A peptide-containing protein inhibits the growth of cells compromised for release factor activity and leads to errors in stop codon recognition. We propose that the nascent 2A peptide interacts with ribosomes to drive a highly unusual and specific "termination" reaction, despite the presence of a proline codon in the A site. After this, the majority of ribosomes continue translation, generating the separate downstream product.
Figures
FIG. 1.
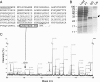
Identification of the C-terminal amino acid of the upstream product as glycine 18. (A) Amino acid sequence of the expected upstream reaction product released from TAP-DHFR-2A-pL by the 2A reaction and tobacco etch virus protease up to the end of 2A plus, in parentheses, the first two downstream amino acids. The identified C-terminal peptide is boxed, and the corresponding peak in the mass spectrum is indicated (C). Underlined sequences were not represented in the mass spectrum. (B) Fractions from the purification (see Materials and Methods) were resolved by SDS-PAGE and stained with Coomassie blue. Positions of marker proteins are marked on the left (molecular sizes are in kilodaltons), as is the purified upstream product of the 2A reaction (arrow). Lanes are whole-cell extract (Load), material not bound to the IgG resin (FT), and eluates from the IgG resin (IgG) and calmodulin resin (Cal). No extension product (predicted molecular mass, 42 kDa) was seen, and Western blotting confirmed that the 2A reaction is very efficient in this construct (data not shown). (C) Partial MALDI-TOF mass spectrum of peptides derived by in-gel tryptic digestion of purified material cut from a gel similar to that shown in panel B. The C-terminal peptide peak is indicated (black arrow). Positions where peptides ending in proline 17, glycine 18, or the downstream arginine (R20) would have been seen are indicated with gray arrows.
FIG. 2.
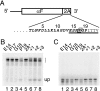
The 2A reaction proceeds with the final codons that encode 2A in the peptidyltransferase center. (A) Outline of construct and 2A sequence. The construct comprises an abbreviated S. cerevisiae pro-α-factor, followed by sequences that encode the FMDV 2A peptide. PCR was used to generate transcription templates from this ending with nucleotides that encode different amino acids from glutamate at position 14 of 2A to proline 19, and also additional amino acids (one, two, three, or four) downstream of proline 19 (underlined). An additional template ended at proline 19 but included an inactivating proline 17 (boxed) mutation to alanine (2A*). (B) In vitro translation reaction mixtures prepared with wheat germ lysate and [35S]methionine with equal amounts of the mRNAs from panel A. Reaction products were subjected to NuPAGE (Invitrogen) SDS-PAGE and visualized with a PhosphorImager. The position of the released upstream product is marked (up), as is that of tRNA adducts (brace). (C) Same as panel B, but samples were treated with RNase prior to resolution on the gel.
FIG. 3.
Toeprinting identifies a ribosomal pause at 2A. (A) Outline of constructs. These contain abbreviated S. cerevisiae pro-α-factor and bovine prolactin sequences flanking amino acids corresponding to the 2A peptide and the CPA1 uORF (AAP) (bold, italics) (see Materials and Methods for details). The proline changed to alanine in 2A* is boxed as in Fig. 1. (B) Toeprinting reactions were carried out on translation reactions initiated on the indicated transcripts with primer ppLanti (see Materials and Methods). Where indicated, arginine (Arg) and/or cycloheximide (Cyh) were added to the reaction mixtures. Cycloheximide was added at the beginning of the reaction (no-translation control) or after 10 min. Controls with no template (−RNA) or extract (−EXT) are shown. Positions of specific pauses on mRNA are marked as follows: initiator codon, open arrow; AAP, brace; 2A, filled arrow. Dideoxynucleotide sequencing reactions with primer ppLanti were run alongside the toeprinting reactions. The mutation that yields the proline-alanine change is marked (arrowhead). The nucleotide complementary to the dideoxynucleotide added to each sequencing reaction mixture is indicated below the corresponding lane so that the sequence of the template can be directly deduced; the 5′to-3′ sequence reads from top to bottom. The relevant portion of the 2A sequence is shown to the right as a series of triplets corresponding to codons in the mRNA. The glycine 18 codon is boxed, and asterisks mark the positions of the 2A-specific toeprints.
FIG. 4.
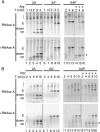
Rapid release of the upstream 2A reaction product from tRNA. (A) In vitro translation reactions were carried out with N. crassa translation extract and [35S]methionine, and equal amounts of synthetic capped, polyadenylated transcripts that encode the constructs indicated (as in Fig. 3A) were run on NuPAGE gels (Invitrogen) and visualized by autoradiography. The extension (E) and separated upstream (up) and downstream (down) 2A reaction products are indicated, as are tRNA adducts (brace) and the AAP-paused species (*). Reactions including the AAP template were carried out at either a low (10 μM) or a high (500 μM) arginine concentration. Samples were taken at the times indicated. Edeine was added to 1 mM at 2 min to inhibit further initiation. Note that the upstream product is degraded over time in the N. crassa extracts. (B) Same as panel A, except that translations were carried out with wheat germ extract. Samples were taken at the times indicated, and edeine was added at 1 min.
FIG. 5.
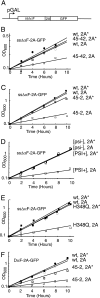
Cells limited in eRF1 activity are sensitive to the overexpression of a 2A-containing protein. (A) ssΔαF-2A-GFP reporter. Transcription is driven by the strong GAL1 promoter. (B) Isogenic wild-type (wt; circles) and _sup45_-42 mutant (45-42; squares) cells were transformed with plasmids that encode ssΔαF-2A-GFP (2A; open symbols) or ssΔαF-2A*-GFP (2A*; closed symbols), grown to mid-log phase at 30°C in 2% raffinose medium, and transferred into medium containing 1% (wt/vol) each galactose and raffinose to induce transcription from pGAL1. The growth of the cultures, which were diluted to maintain conditions suitable for logarithmic growth, was monitored at 30°C as the relative OD600 over time (in hours). (C) Same as panel B but with isogenic wild-type (wt; circles) or _sup45_-2 (45-2; squares) cells, which were grown at 34°C, the semipermissive temperature for the _sup45_-2 mutant cells. (D) Same as panel B but with isogenic [psi−] and strong [PSI+] strains. (E) Same as panel B, except that isogenic wild-type eRF3 and H348Q mutant eRF3 were used. (F) Same as panel B, but cells were transformed with modified reporters containing an N-terminal DAP2 signal sequence which directs cotranslational translocation into the endoplasmic reticulum.
FIG. 6.
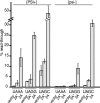
Expression of 2A leads to increased stop codon suppression in trans. [psi−] and strong [PSI+] strains were transformed with plasmids that encode _lacZ_-_STOP_-luc reporters containing the indicated stop codon (plus the downstream nucleotide) along with an empty vector (vector) or a plasmid expressing ssΔαF-2A-GFP (2A) or ssΔαF-2A*-GFP (2A*). These were grown, expression of 2A/2A* was induced, and readthrough assays were carried out (see Materials and Methods). Readthrough is presented as a percentage of the reading obtained with a sense control with a GCA (alanine) codon in place of the stop codon. Each bar is the average measurement from three or more independent transformants with standard deviations indicated.
FIG. 7.
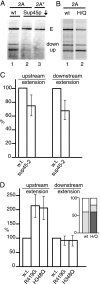
The outcome of ribosomal transit through the 2A coding sequences is altered by changes in RF activity. (A, B) In vitro translation reaction mixtures assembled with mRNA templates including 2A and 2A* as in Fig. 2 but with S. cerevisiae translation extract. Reaction mixtures were assembled with extract from wild-type (wt) or Sup45p-depleted (Sup45p↓) cells (A) and cells expressing wild-type Sup35p or the H438Q (H/Q) variant of Sup35p (B) (42). Reaction products are labeled as in Fig. 4. (C) Anti-2A and anti-GFP antibodies were used to isolate extension and either upstream or downstream 2A reaction products from lysates of isogenic wild-type and _sup45_-2 mutant cells expressing ssΔαF-2A-GFP and pulse-labeled with [35S]methionine following growth at 37°C for 1 h (see Materials and Methods and Results). Immunoprecipitated products were resolved by SDS-PAGE and quantified, and the ratio of the upstream or downstream product to the extension product was determined. Results of three independent experiments were averaged, in each case setting the value obtained for wild-type cells to 100% and expressing that for _sup45_-2 mutant cells relative to this, with 1 standard deviation indicated. To test the significance of these data, an unpaired, two-tailed t test was carried out on the initial data sets, yielding P values of 0.03 and 0.04, respectively, for the means of the data for upstream/elongation and downstream/elongation ratios from the two strains being the same. (D) Same as panel C, except that strains were the isogenic wild type and the R419G and H348Q mutants. In the inset, data for a single experiment are shown as the proportion of each of the different translational outcomes—extension (clear box), complete 2A reaction (light gray), and ribosomal drop off at 2A (dark gray).
FIG. 8.
Model for translation termination at 2A. See text for details.
Similar articles
- Orchestrating ribosomal activity from inside: effects of the nascent chain on the peptidyltransferase centre.
Yan F, Doronina VA, Sharma P, Brown JD. Yan F, et al. Biochem Soc Trans. 2010 Dec;38(6):1576-80. doi: 10.1042/BST0381576. Biochem Soc Trans. 2010. PMID: 21118129 Review. - Misdecoding of rare CGA codon by translation termination factors, eRF1/eRF3, suggests novel class of ribosome rescue pathway in S. cerevisiae.
Wada M, Ito K. Wada M, et al. FEBS J. 2019 Feb;286(4):788-802. doi: 10.1111/febs.14709. Epub 2018 Dec 11. FEBS J. 2019. PMID: 30471181 Free PMC article. - Inhibition of translation termination mediated by an interaction of eukaryotic release factor 1 with a nascent peptidyl-tRNA.
Janzen DM, Frolova L, Geballe AP. Janzen DM, et al. Mol Cell Biol. 2002 Dec;22(24):8562-70. doi: 10.1128/MCB.22.24.8562-8570.2002. Mol Cell Biol. 2002. PMID: 12446775 Free PMC article. - Eukaryotic release factor 3 is required for multiple turnovers of peptide release catalysis by eukaryotic release factor 1.
Eyler DE, Wehner KA, Green R. Eyler DE, et al. J Biol Chem. 2013 Oct 11;288(41):29530-8. doi: 10.1074/jbc.M113.487090. Epub 2013 Aug 20. J Biol Chem. 2013. PMID: 23963452 Free PMC article. - Structural aspects of translation termination on the ribosome.
Korostelev AA. Korostelev AA. RNA. 2011 Aug;17(8):1409-21. doi: 10.1261/rna.2733411. Epub 2011 Jun 23. RNA. 2011. PMID: 21700725 Free PMC article. Review.
Cited by
- A pan-cancer analysis reveals nonstop extension mutations causing SMAD4 tumour suppressor degradation.
Dhamija S, Yang CM, Seiler J, Myacheva K, Caudron-Herger M, Wieland A, Abdelkarim M, Sharma Y, Riester M, Groß M, Maurer J, Diederichs S. Dhamija S, et al. Nat Cell Biol. 2020 Aug;22(8):999-1010. doi: 10.1038/s41556-020-0551-7. Epub 2020 Jul 27. Nat Cell Biol. 2020. PMID: 32719554 - Effects of different 2A peptides on transgene expression mediated by tricistronic vectors in transfected CHO cells.
Li YM, Wang M, Wang TY, Wei YG, Guo X, Mi CL, Zhao CP, Cao XX, Dou YY. Li YM, et al. Mol Biol Rep. 2020 Jan;47(1):469-475. doi: 10.1007/s11033-019-05153-3. Epub 2019 Oct 28. Mol Biol Rep. 2020. PMID: 31659692 - An atypical unfolded protein response in heat shocked cells.
Heldens L, Hensen SM, Onnekink C, van Genesen ST, Dirks RP, Lubsen NH. Heldens L, et al. PLoS One. 2011;6(8):e23512. doi: 10.1371/journal.pone.0023512. Epub 2011 Aug 10. PLoS One. 2011. PMID: 21853144 Free PMC article. - Detecting gene expression in Caenorhabditis elegans.
Calarco JA, Taylor SR, Miller DM 3rd. Calarco JA, et al. Genetics. 2025 Jan 8;229(1):1-108. doi: 10.1093/genetics/iyae167. Genetics. 2025. PMID: 39693264 Free PMC article. Review. - Non-canonical translation in RNA viruses.
Firth AE, Brierley I. Firth AE, et al. J Gen Virol. 2012 Jul;93(Pt 7):1385-1409. doi: 10.1099/vir.0.042499-0. Epub 2012 Apr 25. J Gen Virol. 2012. PMID: 22535777 Free PMC article. Review.
References
- Alkalaeva, E. Z., A. V. Pisarev, L. Y. Frolova, L. L. Kisselev, and T. V. Pestova. 2006. In vitro reconstitution of eukaryotic translation reveals cooperativity between release factors eRF1 and eRF3. Cell 1251125-1136. - PubMed
- Anthony, D. D., and W. C. Merrick. 1992. Analysis of 40 S and 80 S complexes with mRNA as measured by sucrose density gradients and primer extension inhibition. J. Biol. Chem. 2671554-1562. - PubMed
- Atkins, J. F., N. M. Wills, G. Loughran, C. Y. Wu, K. Parsawar, M. D. Ryan, C. H. Wang, and C. C. Nelson. 2007. A case for “StopGo”: reprogramming translation to augment codon meaning of GGN by promoting unconventional termination (Stop) after addition of glycine and then allowing continued translation (Go). RNA 13803-810. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- BB/E/01070911/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- G117/395/MRC_/Medical Research Council/United Kingdom
- R01 GM047498/GM/NIGMS NIH HHS/United States
- C20035/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- GM47498/GM/NIGMS NIH HHS/United States
- BB/E009093/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/E009093/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/E010709/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- C20418/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases