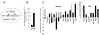ER71 acts downstream of BMP, Notch, and Wnt signaling in blood and vessel progenitor specification - PubMed (original) (raw)
. 2008 May 8;2(5):497-507.
doi: 10.1016/j.stem.2008.03.008.
Changwon Park, Ho Lee, Jesse J Lugus, Seok Hyung Kim, Elizabeth Arentson, Yun Shin Chung, Gustavo Gomez, Michael Kyba, Shuo Lin, Ralf Janknecht, Dae-Sik Lim, Kyunghee Choi
Affiliations
- PMID: 18462699
- PMCID: PMC2683414
- DOI: 10.1016/j.stem.2008.03.008
ER71 acts downstream of BMP, Notch, and Wnt signaling in blood and vessel progenitor specification
Dongjun Lee et al. Cell Stem Cell. 2008.
Abstract
FLK1-expressing (FLK1(+)) mesoderm generates blood and vessels. Here, we show that combined BMP, Notch, and Wnt signaling is necessary for efficient FLK1(+) mesoderm formation from embryonic stem cells (ESCs). Inhibition of BMP, Notch, and Wnt signaling pathways greatly decreased the generation of FLK1(+) mesoderm and expression of the Ets transcription factor Er71. Enforced expression of ER71 in ESCs resulted in a robust induction of FLK1(+) mesoderm; rescued the generation of FLK1(+) mesoderm when blocked by BMP, Notch, and Wnt inhibition; and enhanced hematopoietic and endothelial cell generation. Er71-deficient mice had greatly reduced FLK1 expression, died early in gestation, and displayed severe blood and vessel defects that are highly reminiscent of the Flk1 null mouse phenotype. Collectively, we provide compelling evidence that ER71 functions downstream of BMP, Notch, and Wnt signals and regulates FLK1(+) mesoderm, blood, and vessel development.
Figures
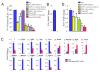
Figure 1. BMP, Notch and Wnt inhibition blocks the generation of FLK1+ cells and abrogates Er71 expression during ES cell differentiation
ES cells were differentiated in serum (FCS), inhibitors were added on day 1, and day 3 EB cells were analyzed for FLK1 expression (A) and subjected to gene expression (B–D). *P<0.05 versus FCS. Genes were normalized against Gapdh and then the ratio of the gene quantity (FCS) to gene quantity (FCS + inhibitors) was determined to yield fold change shown on the Y-axis.
Figure 2. Er71 is expressed transiently in early embryos and differentiating ES cells
(A) In situ hybridization analysis of Er71. PS, primitive streak; Bl, blood islands; Al, allantois; Am, amnion; A, anterior; P, posterior; D, dorsal; V, ventral (B, C) Er71 expression analysis. RNA obtained from differentiating ES cells (B) or various cell populations sorted from day 2.75 EBs (C) was used to examine Er71 expression. The value normalized against Gapdh is shown on the Y-axis.
Figure 3. ER71 can induce FLK1+ mesoderm by activating Flk1 expression
(A) Schematic diagram of the inducible ER71 (iER71) ES cells used, with indicated loci carrying alterations allowing for production of the rtTA, expression of the Er71 cDNA and generation of hCD4 as a surrogate marker for Scl. (B, C) Er71 induction upon Dox treatment and the ER71-mediated transcriptional program. iER71 ES cells were differentiated for 1 day in serum or for 2 days in SR and then treated with 1 μg/ml Dox for an additional 2 days. RNA was prepared and used for qRT-PCR. Genes were normalized against Gapdh and the ratio of the gene quantity (+Dox) to gene quantity (−Dox) was determined to yield normalized fold change. (D) iER71 ES cells were differentiated as described. BMP4 was added on day 2. The resulting cells were FACS analyzed for FLK1. Numbers in insets indicate the percentage of FLK1+ cells. (E) iER71 ES cells were differentiated in serum as described. Noggin, DAPT and DKK1, singularly or in combination, were added on day 1 of differentiation. Dox was added on day 1 and FLK1+ cells were analyzed on day 3. Numbers in insets indicate the percentage of FLK1+ cells. (F) (upper) Schematic diagram of the Flk1 promoter used for luciferase assay. Diamonds indicate potential Ets binding sites (red in sense and blue in anti-sense). For mutagenesis, potential Ets sites GGAA/T (sense) and A/TTCC (anti-sense) were mutated to TTAA/T and A/TTTT, respectively. (lower) 293T cells were transfected with pGL3 or pGL3-Flk1 promoter-luciferase reporter plasmid with or without pCS3-Myc-Er71(WT or MT). Fire fly luciferase activity was normalized by Renilla luciferase activity. (G) iER71 ES cells differentiated in serum in the presence of Dox were cross-linked, sonicated and subjected to ChIP assay. Numbers on the X-axis indicate the locations of amplicons of each qPCR primer set on Flk1. The value normalized against IgG IP values is shown on the Y-axis.
Figure 3. ER71 can induce FLK1+ mesoderm by activating Flk1 expression
(A) Schematic diagram of the inducible ER71 (iER71) ES cells used, with indicated loci carrying alterations allowing for production of the rtTA, expression of the Er71 cDNA and generation of hCD4 as a surrogate marker for Scl. (B, C) Er71 induction upon Dox treatment and the ER71-mediated transcriptional program. iER71 ES cells were differentiated for 1 day in serum or for 2 days in SR and then treated with 1 μg/ml Dox for an additional 2 days. RNA was prepared and used for qRT-PCR. Genes were normalized against Gapdh and the ratio of the gene quantity (+Dox) to gene quantity (−Dox) was determined to yield normalized fold change. (D) iER71 ES cells were differentiated as described. BMP4 was added on day 2. The resulting cells were FACS analyzed for FLK1. Numbers in insets indicate the percentage of FLK1+ cells. (E) iER71 ES cells were differentiated in serum as described. Noggin, DAPT and DKK1, singularly or in combination, were added on day 1 of differentiation. Dox was added on day 1 and FLK1+ cells were analyzed on day 3. Numbers in insets indicate the percentage of FLK1+ cells. (F) (upper) Schematic diagram of the Flk1 promoter used for luciferase assay. Diamonds indicate potential Ets binding sites (red in sense and blue in anti-sense). For mutagenesis, potential Ets sites GGAA/T (sense) and A/TTCC (anti-sense) were mutated to TTAA/T and A/TTTT, respectively. (lower) 293T cells were transfected with pGL3 or pGL3-Flk1 promoter-luciferase reporter plasmid with or without pCS3-Myc-Er71(WT or MT). Fire fly luciferase activity was normalized by Renilla luciferase activity. (G) iER71 ES cells differentiated in serum in the presence of Dox were cross-linked, sonicated and subjected to ChIP assay. Numbers on the X-axis indicate the locations of amplicons of each qPCR primer set on Flk1. The value normalized against IgG IP values is shown on the Y-axis.
Figure 4. ER71 can induce hematopoietic and endothelial cell generation
(A) iER71 ES cells were differentiated in SR or serum and Dox (1 μg/ml) was added on day 3. BMP4 and VEGFA165 were added on day 2 and day 3, respectively. In all cases, cells were FACS analyzed for FLK1, hCD4 or VE-Cadherin (VE-Cad) expression on day 5 (FCS) or day 6 (SR). (left panels) Numbers in a given box indicate the percentage of cells that are FLK1−hCD4− (lower left) FLK1+hCD4− (lower right), FLK1+hCD4+ (upper right), or FLK1−hCD4+ (upper left). (right panels) Numbers in a given box indicate the percentage of VE-Cad+ cells. (B) iER71 cells differentiated for 6 days in serum free conditions with indicated factors were replated in methylcellulose medium containing hematopoietic cytokines. Colonies were counted 5–7 days after replating. EB cells generated in SR alone gave rise to few erythoid colonies (average=10.7) with no Mac or E/M colonies. Error bars indicate standard deviations from three independent experiments. Ery; Erythrocytes, Mac; Macrophages, E/M; Erythroid/Macrophage colonies. (C) Transient expression of ER71 increases the generation of primitive erythroid progenitors. Dox was added to the culture at the indicated day and kept until day 4 or washed out (WO) the next day. On day 4, iER71 EB cells were replated for primitive erythroid colonies. Colonies were counted 4 days later. Data represent four independent experiments. (D) Hematopoietic and endothelial cell development upon ER71 overexpression using embryo-derived cell culture. Pooled embryos between E8.0–E8.5 were dissociated into single cells and infected with MSCV or MSCV-ER71-IRES-GFP virus. Subsequently, cells cultured on OP9 cells were subjected to hematopoietic colony replating (upper, values are mean±s.e.m. from two independent experiments) or PECAM1 staining (lower, representative images are shown).
Figure 5. Disruption of the murine Er71 locus
(A) Structures of the mouse Er71 gene, the pGKpuro targeting vector, and the targeted allele after homologous recombination. Black boxes depict exons. The flanking genomic probes used for Southern blot analysis are indicated with gray squares. Only the EcoRI restriction enzyme site relevant to the targeting construct and screening strategies is shown. (B) Southern blot analyses of representative Er71+/− ES cell clones and genotyping of ES cells and E9.5 embryos by PCR. The wild type (15.8kb) and the mutant allele (8.5kb) detected with the 5′ external probe as well as the wild type (15.8kb) and the mutant allele (6.8kb) detected with the 3′ external probe after EcoRI digest are shown. PCR products of the wild type Er71 allele (primer 1, 2: 1000 bp) and the mutant allele (primer 1, 3: 700 bp) are shown. (C) Gross morphology of _Er71_−/− embryos at E9.5. (a, c, embryo proper; b, d, yolk sac). Scale bars; 200 μm. (D) Gene expression analysis. RNA was prepared from E8.5 (n=2) embryos and subjected to qRT-PCR. The value normalized against Gapdh is shown on the Y-axis.
Figure 6. Histology of Er71 mutants
(A, B) H & E staining of E8.5 embryos. The boxed area is shown at higher magnification. Bl, blood island; DA, dorsal aorta. Scale bars; 50μm. (C) Sagittal sections of wild type and _Er71_−/− hearts from E9.5 embryos. Ec, endocardium; Mc, myocardium. Scale bars; 50 μm. (D) Sagittal sections of the placenta of E9.5 embryos. Higher magnification of the labyrinth layers of the wild type and _Er71_−/− placentas is shown in the box. dec, maternal decidual tissue; sp, spongiotrophoblast layer; la, labyrinth layer. Scale bars; 200 μm.
Figure 7. Er71_−/_− embryos show defects in blood and vessel development
(A) (left) Benzidine staining. Scale bars; 200 μm. (right) Hematopoietic progenitor assay. Cells prepared from E8.5 YS were replated into methycellulose containing hematopoietic cytokines. (B) FLK1 expression in E7.5 embryos. FLK1 is expressed in primitive streak (PS), blood islands (Bl), allantois (Al) and amnion (Am) in the wild type but not in the mutants. Scale bars; 50 μm. (C) FLK1 expression in E8.5 embryos. (upper) Whole mount FLK1 staining of the embryo proper. Scale bars; 200 μm. (lower) Immunofluorescent FLK1 (green) and PECAM1 (red) staining of the yolk sac. Nuclei were stained with DAPI (blue). Em, embryo proper. Scale bars; 50 μm. (D) Whole-mount PECAM1 or FLK1 staining of E9.5 embryos. The boxed area is the vasculature of the yolk sac, brain, heart and intersomitic regions at higher magnification. Scale bars; 200 μm.
Similar articles
- A hierarchical order of factors in the generation of FLK1- and SCL-expressing hematopoietic and endothelial progenitors from embryonic stem cells.
Park C, Afrikanova I, Chung YS, Zhang WJ, Arentson E, Fong Gh Gh, Rosendahl A, Choi K. Park C, et al. Development. 2004 Jun;131(11):2749-62. doi: 10.1242/dev.01130. Development. 2004. PMID: 15148304 - ER71 specifies Flk-1+ hemangiogenic mesoderm by inhibiting cardiac mesoderm and Wnt signaling.
Liu F, Kang I, Park C, Chang LW, Wang W, Lee D, Lim DS, Vittet D, Nerbonne JM, Choi K. Liu F, et al. Blood. 2012 Apr 5;119(14):3295-305. doi: 10.1182/blood-2012-01-403766. Epub 2012 Feb 17. Blood. 2012. PMID: 22343916 Free PMC article. - Wnt, activin, and BMP signaling regulate distinct stages in the developmental pathway from embryonic stem cells to blood.
Nostro MC, Cheng X, Keller GM, Gadue P. Nostro MC, et al. Cell Stem Cell. 2008 Jan 10;2(1):60-71. doi: 10.1016/j.stem.2007.10.011. Cell Stem Cell. 2008. PMID: 18371422 Free PMC article. - ETS Transcription Factor ETV2/ER71/Etsrp in Hematopoietic and Vascular Development.
Sumanas S, Choi K. Sumanas S, et al. Curr Top Dev Biol. 2016;118:77-111. doi: 10.1016/bs.ctdb.2016.01.005. Epub 2016 Feb 12. Curr Top Dev Biol. 2016. PMID: 27137655 Review. - Cell fate decisions in early blood vessel formation.
Ema M, Rossant J. Ema M, et al. Trends Cardiovasc Med. 2003 Aug;13(6):254-9. doi: 10.1016/s1050-1738(03)00105-1. Trends Cardiovasc Med. 2003. PMID: 12922023 Review.
Cited by
- Etv2 transcriptionally regulates Yes1 and promotes cell proliferation during embryogenesis.
Singh BN, Gong W, Das S, Theisen JWM, Sierra-Pagan JE, Yannopoulos D, Skie E, Shah P, Garry MG, Garry DJ. Singh BN, et al. Sci Rep. 2019 Jul 5;9(1):9736. doi: 10.1038/s41598-019-45841-5. Sci Rep. 2019. PMID: 31278282 Free PMC article. - Ferric citrate and apo-transferrin enable erythroblast maturation with β-globin from hemogenic endothelium.
Jeon SB, Koh H, Han AR, Kim J, Lee S, Lee JH, Im SS, Yoon YS, Lee JH, Lee JY. Jeon SB, et al. NPJ Regen Med. 2023 Aug 25;8(1):46. doi: 10.1038/s41536-023-00320-4. NPJ Regen Med. 2023. PMID: 37626061 Free PMC article. - Feedback Mechanisms Regulate Ets Variant 2 (Etv2) Gene Expression and Hematoendothelial Lineages.
Koyano-Nakagawa N, Shi X, Rasmussen TL, Das S, Walter CA, Garry DJ. Koyano-Nakagawa N, et al. J Biol Chem. 2015 Nov 20;290(47):28107-28119. doi: 10.1074/jbc.M115.662197. Epub 2015 Sep 22. J Biol Chem. 2015. PMID: 26396195 Free PMC article. - Pioneer factor ETV2 safeguards endothelial cell specification by recruiting the repressor REST to restrict alternative lineage commitment.
Chen D, Fan X, Sun N, Wang K, Gong L, Melero-Martin JM, Pu WT. Chen D, et al. Nat Cardiovasc Res. 2025 Jun;4(6):689-709. doi: 10.1038/s44161-025-00660-y. Epub 2025 Jun 10. Nat Cardiovasc Res. 2025. PMID: 40495012 - VEGF/Flk1 signaling cascade transactivates Etv2 gene expression.
Rasmussen TL, Shi X, Wallis A, Kweon J, Zirbes KM, Koyano-Nakagawa N, Garry DJ. Rasmussen TL, et al. PLoS One. 2012;7(11):e50103. doi: 10.1371/journal.pone.0050103. Epub 2012 Nov 19. PLoS One. 2012. PMID: 23185546 Free PMC article.
References
- Bartel FO, Higuchi T, Spyropoulos DD. Mouse models in the study of the Ets family of transcription factors. Oncogene. 2000;19:6443–6454. - PubMed
- Brown TA, McKnight SL. Specificities of protein-protein and protein-DNA interaction of GABP alpha and two newly defined ets-related proteins. Genes Dev. 1992;6:2502–2512. - PubMed
- Carmeliet P, Ferreira V, Breier G, Pollefeyt S, Kieckens L, Gertsenstein M, Fahrig M, Vandenhoeck A, Harpal K, Eberhardt C, Declercq C, Pawling J, Moons L, Collen D, Risau W, Nagy A. Abnormal blood vessel development and lethality in embryos lacking a single VEGF allele. Nature. 1996;380:435–439. - PubMed
- Chung YS, Zhang WJ, Arentson E, Kingsley PD, Palis J, Choi K. Lineage analysis of the hemangioblast as defined by FLK1 and SCL expression. Development. 2002;129:5511–5520. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 HL081186/HL/NHLBI NIH HHS/United States
- Z01 DK054508/ImNIH/Intramural NIH HHS/United States
- R01 HL055337/HL/NHLBI NIH HHS/United States
- R01 DK054508/DK/NIDDK NIH HHS/United States
- R01 HL063736/HL/NHLBI NIH HHS/United States
- DK54508/DK/NIDDK NIH HHS/United States
- R29 HL055337/HL/NHLBI NIH HHS/United States
- HL55337/HL/NHLBI NIH HHS/United States
- HL63736/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases