Bird-like sex chromosomes of platypus imply recent origin of mammal sex chromosomes - PubMed (original) (raw)
doi: 10.1101/gr.7101908. Epub 2008 May 7.
Paul D Waters, Pat Miethke, Willem Rens, Daniel McMillan, Amber E Alsop, Frank Grützner, Janine E Deakin, Camilla M Whittington, Kyriena Schatzkamer, Colin L Kremitzki, Tina Graves, Malcolm A Ferguson-Smith, Wes Warren, Jennifer A Marshall Graves
Affiliations
- PMID: 18463302
- PMCID: PMC2413164
- DOI: 10.1101/gr.7101908
Bird-like sex chromosomes of platypus imply recent origin of mammal sex chromosomes
Frédéric Veyrunes et al. Genome Res. 2008 Jun.
Abstract
In therian mammals (placentals and marsupials), sex is determined by an XX female: XY male system, in which a gene (SRY) on the Y affects male determination. There is no equivalent in other amniotes, although some taxa (notably birds and snakes) have differentiated sex chromosomes. Birds have a ZW female: ZZ male system with no homology with mammal sex chromosomes, in which dosage of a Z-borne gene (possibly DMRT1) affects male determination. As the most basal mammal group, the egg-laying monotremes are ideal for determining how the therian XY system evolved. The platypus has an extraordinary sex chromosome complex, in which five X and five Y chromosomes pair in a translocation chain of alternating X and Y chromosomes. We used physical mapping to identify genes on the pairing regions between adjacent X and Y chromosomes. Most significantly, comparative mapping shows that, contrary to earlier reports, there is no homology between the platypus and therian X chromosomes. Orthologs of genes in the conserved region of the human X (including SOX3, the gene from which SRY evolved) all map to platypus chromosome 6, which therefore represents the ancestral autosome from which the therian X and Y pair derived. Rather, the platypus X chromosomes have substantial homology with the bird Z chromosome (including DMRT1) and to segments syntenic with this region in the human genome. Thus, platypus sex chromosomes have strong homology with bird, but not to therian sex chromosomes, implying that the therian X and Y chromosomes (and the SRY gene) evolved from an autosomal pair after the divergence of monotremes only 166 million years ago. Therefore, the therian X and Y are more than 145 million years younger than previously thought.
Figures
Figure 1.
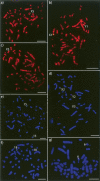
BACs mapped by FISH to (a–d) the sex chromosomes and (e–g) autosomes orthologous to the human X on male platypus metaphase spreads: (a) CH236-72 A21; (b) CH236-3 C11; (c) CH236-20 M7; (d) (green) CH236-271 I19 and (red) CH236-165 F5; (e) (green) CH236-359 L11 (representing UltraContig462) and (red) CH236-97 I3 (chromosome 15 anchor BAC); (f) (green) CH236-427 K7 (representing UltraContig222) and (red) Ch236-330 L7 (chromosome 18 anchor BAC): (g) (green) CH236-337 O23 (representing UltraContig420) and (red) CH236-30 G14 (representing UltraContig519). Scale bars, 10 μm.
Figure 2.
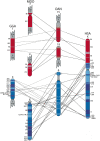
The 10 sex chromosomes from male platypus with the location of the 32 BACs mapped in this study and four BACs/genes mapped in recent studies (colored bands). BAC numbers are indicated, and homology with chicken is represented by different colors. BACs that hybridized to both an X and a Y revealed pseudoautosomal regions and are connected by lines.
Figure 3.
The human (HSA) X chromosome compared to orthologous platypus (OAN), opossum (MDO), and chicken (GGA) chromosomes. All shades of blue are orthologous to the X conserved region (XCR) of the placental mammal X chromosome. Royal blue on the human X, opossum X, and chicken 4p were anchored to platypus chromosome 6 prior to this study; navy blue regions on these chromosomes are orthologous to unanchored UltraContigs in the platypus assembly; whereas pale blue is predicted orthology with platypus chromosome 6. Red is orthologous to X added region (XAR), and pink is the predicted orthology to platypus chromosomes 15 and 18. None of the X added orthologous regions were anchored in platypus before this study. The numbers of all unanchored UltraContigs (see Table 1 for list of anchoring BACs) are listed left of the chicken and opossum chromosomes, and right of the human X. FISH locations of BACs on platypus chromosomes 6, 15, and 18 are shown, and lines indicate orthology with the human, opossum, and chicken genomes. Some of the platypus UltraContigs have been broken up in human, opossum, and/or chicken. Vertical lines next to platypus chromosome 6 indicate that FISH could not achieve resolution between BACs.
Figure 4.
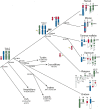
Phylogeny displaying the different sex chromosome systems of major vertebrate groups. Homology between genomes, and with ancestral autosomes, is indicated by the same color. Key events are marked on the tree. (Green) A chicken-like ZW system appears to be ancestral to all amniotes, which was retained in birds and caught up in the meiotic chain of monotremes. There was a switch from ZW to XY systems independently in both therian and monotreme mammals. (Red) The region added to the sex chromosomes of placental mammals was broken independently in opossum and monotremes. (Green) The autosome orthologous to the bird Z chromosome was broken in the therian ancestor and subsequently scrambled onto several autosomes in mouse; therefore only the chromosome carrying Dmrt1 is displayed in that species.
Similar articles
- The multiple sex chromosomes of platypus and echidna are not completely identical and several share homology with the avian Z.
Rens W, O'Brien PC, Grützner F, Clarke O, Graphodatskaya D, Tsend-Ayush E, Trifonov VA, Skelton H, Wallis MC, Johnston S, Veyrunes F, Graves JA, Ferguson-Smith MA. Rens W, et al. Genome Biol. 2007;8(11):R243. doi: 10.1186/gb-2007-8-11-r243. Genome Biol. 2007. PMID: 18021405 Free PMC article. - Sex determination in mammals--before and after the evolution of SRY.
Wallis MC, Waters PD, Graves JA. Wallis MC, et al. Cell Mol Life Sci. 2008 Oct;65(20):3182-95. doi: 10.1007/s00018-008-8109-z. Cell Mol Life Sci. 2008. PMID: 18581056 Free PMC article. Review. - Sex determination in platypus and echidna: autosomal location of SOX3 confirms the absence of SRY from monotremes.
Wallis MC, Waters PD, Delbridge ML, Kirby PJ, Pask AJ, Grützner F, Rens W, Ferguson-Smith MA, Graves JA. Wallis MC, et al. Chromosome Res. 2007;15(8):949-59. doi: 10.1007/s10577-007-1185-3. Epub 2008 Jan 9. Chromosome Res. 2007. PMID: 18185981 - The status of dosage compensation in the multiple X chromosomes of the platypus.
Deakin JE, Hore TA, Koina E, Marshall Graves JA. Deakin JE, et al. PLoS Genet. 2008 Jul 25;4(7):e1000140. doi: 10.1371/journal.pgen.1000140. PLoS Genet. 2008. PMID: 18654631 Free PMC article. - Mammalian sex--Origin and evolution of the Y chromosome and SRY.
Waters PD, Wallis MC, Marshall Graves JA. Waters PD, et al. Semin Cell Dev Biol. 2007 Jun;18(3):389-400. doi: 10.1016/j.semcdb.2007.02.007. Epub 2007 Feb 24. Semin Cell Dev Biol. 2007. PMID: 17400006 Review.
Cited by
- Advances and challenges in genetic technologies to produce single-sex litters.
Douglas C, Turner JMA. Douglas C, et al. PLoS Genet. 2020 Jul 23;16(7):e1008898. doi: 10.1371/journal.pgen.1008898. eCollection 2020 Jul. PLoS Genet. 2020. PMID: 32701961 Free PMC article. Review. - Fragmentation by major dams and implications for the future viability of platypus populations.
Mijangos JL, Bino G, Hawke T, Kolomyjec SH, Kingsford RT, Sidhu H, Grant T, Day J, Dias KN, Gongora J, Sherwin WB. Mijangos JL, et al. Commun Biol. 2022 Nov 3;5(1):1127. doi: 10.1038/s42003-022-04038-9. Commun Biol. 2022. PMID: 36329312 Free PMC article. - Independent evolution of transcriptional inactivation on sex chromosomes in birds and mammals.
Livernois AM, Waters SA, Deakin JE, Marshall Graves JA, Waters PD. Livernois AM, et al. PLoS Genet. 2013;9(7):e1003635. doi: 10.1371/journal.pgen.1003635. Epub 2013 Jul 18. PLoS Genet. 2013. PMID: 23874231 Free PMC article. - Incomplete transcriptional dosage compensation of chicken and platypus sex chromosomes is balanced by post-transcriptional compensation.
Lister NC, Milton AM, Patel HR, Waters SA, Hanrahan BJ, McIntyre KL, Livernois AM, Horspool WB, Wee LK, Ringel AR, Mundlos S, Robson MI, Shearwin-Whyatt L, Grützner F, Graves JAM, Ruiz-Herrera A, Waters PD. Lister NC, et al. Proc Natl Acad Sci U S A. 2024 Aug 6;121(32):e2322360121. doi: 10.1073/pnas.2322360121. Epub 2024 Jul 29. Proc Natl Acad Sci U S A. 2024. PMID: 39074288 Free PMC article. - Dynamic karyotype evolution and unique sex determination systems in Leptidea wood white butterflies.
Šíchová J, Voleníková A, Dincă V, Nguyen P, Vila R, Sahara K, Marec F. Šíchová J, et al. BMC Evol Biol. 2015 May 19;15:89. doi: 10.1186/s12862-015-0375-4. BMC Evol Biol. 2015. PMID: 25981157 Free PMC article.
References
- Alsop A.E., Miethke P., Rofe R., Koina E., Sankovic N., Deakin J.E., Haines H., Rapkins R.W., Graves J.A.M., Miethke P., Rofe R., Koina E., Sankovic N., Deakin J.E., Haines H., Rapkins R.W., Graves J.A.M., Rofe R., Koina E., Sankovic N., Deakin J.E., Haines H., Rapkins R.W., Graves J.A.M., Koina E., Sankovic N., Deakin J.E., Haines H., Rapkins R.W., Graves J.A.M., Sankovic N., Deakin J.E., Haines H., Rapkins R.W., Graves J.A.M., Deakin J.E., Haines H., Rapkins R.W., Graves J.A.M., Haines H., Rapkins R.W., Graves J.A.M., Rapkins R.W., Graves J.A.M., Graves J.A.M. Characterizing the chromosomes of the Australian model marsupial Macropus eugenii (tammar wallaby) Chromosome Res. 2005;13:627–636. - PubMed
- Bick Y., Sharman G., Sharman G. The chromosomes of the platypus (Ornithorhynchus): Monotremata. Cytobios. 1975;14:17–28.
- Bininda-Emonds O.R.P., Cardillo M., Jones K.E., MacPhee R.D.E., Beck R.M.D., Grenyer R., Price S.A., Vos R.A., Gittleman J.L., Purvis A., Cardillo M., Jones K.E., MacPhee R.D.E., Beck R.M.D., Grenyer R., Price S.A., Vos R.A., Gittleman J.L., Purvis A., Jones K.E., MacPhee R.D.E., Beck R.M.D., Grenyer R., Price S.A., Vos R.A., Gittleman J.L., Purvis A., MacPhee R.D.E., Beck R.M.D., Grenyer R., Price S.A., Vos R.A., Gittleman J.L., Purvis A., Beck R.M.D., Grenyer R., Price S.A., Vos R.A., Gittleman J.L., Purvis A., Grenyer R., Price S.A., Vos R.A., Gittleman J.L., Purvis A., Price S.A., Vos R.A., Gittleman J.L., Purvis A., Vos R.A., Gittleman J.L., Purvis A., Gittleman J.L., Purvis A., Purvis A. The delayed rise of present-day mammals. Nature. 2007;466:507–512. - PubMed
- Charlesworth D., Charlesworth B., Marais G., Charlesworth B., Marais G., Marais G. Steps in the evolution of heteromorphic sex chromosomes. Heredity. 2005;95:118–128. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials