Elucidating the relationship between DISC1, NDEL1 and NDE1 and the risk for schizophrenia: evidence of epistasis and competitive binding - PubMed (original) (raw)
. 2008 Aug 15;17(16):2462-73.
doi: 10.1093/hmg/ddn146. Epub 2008 May 10.
Affiliations
- PMID: 18469341
- PMCID: PMC2486442
- DOI: 10.1093/hmg/ddn146
Elucidating the relationship between DISC1, NDEL1 and NDE1 and the risk for schizophrenia: evidence of epistasis and competitive binding
Katherine E Burdick et al. Hum Mol Genet. 2008.
Abstract
DISC1 influences susceptibility to psychiatric disease and related phenotypes. Intact functions of DISC1 and its binding partners, NDEL1 and NDE1, are critical to neurodevelopmental processes aberrant in schizophrenia (SZ). Despite evidence of an NDEL1-DISC1 protein interaction, there have been no investigations of the NDEL1 gene or the relationship between NDEL1 and DISC1 in SZ. We genotyped six NDEL1 single-nucleotide polymorphisms (SNPs) in 275 Caucasian SZ patients and 200 controls and tested for association and interaction between the functional SNP Ser704Cys in DISC1 and NDEL1. We also evaluated the relationship between NDE1 and DISC1 genotype and SZ. Finally, in a series of in vitro assays, we determined the binding profiles of NDEL1 and NDE1, in relation to DISC1 Ser704Cys. We observed a single haplotype block within NDEL1; the majority of variation was captured by NDEL1 rs1391768. We observed a significant interaction between rs1391768 and DISC1 Ser704Cys, with the effect of NDEL1 on SZ evident only against the background of DISC1 Ser704 homozygosity. Secondary analyses revealed no direct relationship between NDE1 genotype and SZ; however, there was an opposite pattern of risk for NDE1 genotype when conditioned on DISC1 Ser704Cys, with NDE1 rs3784859 imparting a significant effect but only in the context of a Cys-carrying background. In addition, we report opposing binding patterns of NDEL1 and NDE1 to Ser704 versus Cys704, at the same DISC1 binding domain. These data suggest that NDEL1 significantly influences risk for SZ via an interaction with DISC1. We propose a model where NDEL1 and NDE1 compete for binding with DISC1.
Figures
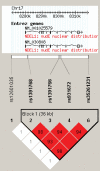
Figure 1.
LD structure of NDEL1 haplotype block. LD (D′) structure using Haploview 3.32 (46). One SNP, rs 2012190, demonstrated extremely rare (<05%) minor allele frequency and was excluded from further analyses. Of the remaining SNPs, four formed a tight haplotype block spanning ∼36 kb, encompassing the NDEL1 gene.
Figure 2.
Effect of NDEL1 rs1391768 genotype in a DISC1 Ser704Cys background. Subjects are grouped by genotype at both NDEL1 rs1391768 and DISC1 Ser704Cys. The _X_-axis represents NDEL1 genotype group. The _Y_-axis represents OR. These data depict the DISC1–NDEL1 genotype interaction graphically. Sample sizes for each group are as follows: NDEL1 G carrier × DISC1 Ser704Ser (SZ = 74, HC = 43); NDEL1 AA × DISC1 Ser704Ser (SZ = 31, HC = 44); NDEL1 G carrier × DISC1 Cys carrier (SZ = 52, HC = 42) and NDEL1 AA × DISC1 Cys carrier (SZ = 83, HC = 65).
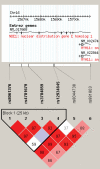
Figure 3.
LD structure of NDE1 haplotype block. LD (D′) structure using Haploview 3.32 (24). Four of the six SNPs formed a tight haplotype block spanning ∼30 kb, within the NDE1 gene.
Figure 4.
Effect of NDE1 rs3784859 genotype in a DISC1 Ser704Cys background. Subjects are grouped by genotype at both NDE1 rs3784859 and DISC1 Ser704Cys. The _X_-axis represents NDE1 genotype group. The _Y_-axis represents OR. These data depict the DISC1–NDE1 genotype interaction graphically. Sample sizes for each group are as follows: NDE1 G carrier × DISC1 Ser704Ser (SZ = 45, HC = 35); NDE1 AA × DISC1 Ser704Ser (SZ = 28, HC = 25); NDE1 G carrier × DISC1 Cys carrier (SZ = 75, HC = 41) and NDE1 AA × DISC1 Cys carrier (SZ = 22, HC = 24).
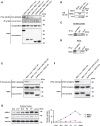
Figure 5.
Protein interaction of DISC1, NDEL1 and NDE1. (A) Self-association of NDEL1 and hetero-oligomerization of NDEL1 and NDE1. HA-tagged DISC1, NDEL1 or GAPDH were transfected with myc-tagged NDEL1 or NDE1 in HEK293 cells. Immunoprecipitates with an anti-HA antibody were analyzed by western blotting with an anti-myc antibody. Myc-tagged NDEL1 is co-precipitated with HA-tagged DISC1 and NDEL1, but not with GAPDH (top panel). NDE1 is also co-precipitated with HA-tagged NDEL1, but not with GAPDH (top panel), suggesting that NDEL1 self-associates and forms a hetero-oligomer with NDE1. For a negative control in co-immunoprecipitation, we used a rabbit IgG and observed no precipitation of target proteins (second panel). The inputs of each protein in co-immunoprecipitation are also shown (bottom panels). (B) The specificities of monoclonal antibodies against NDE1 (see Materials and Methods section) and NDEL1 (24) were evaluated by western blotting. Anti-NDE1 antibody recognizes exogenous NDE1 protein but not exogenous NDEL1 protein. On the other hand, anti-NDEL1 antibody detects exogenous NDEL1 protein but not exogenous NDE1 protein. (C) Endogenous protein interaction of NDE1–DISC1 was confirmed by co-immunoprecipitation with the tissue extract of rat adult cerebral cortex. A monoclonal antibody against NDE1 was used for immunoprecipitation. (D) Endogenous protein interaction of NDE1–NDEL1 was confirmed by co-immunoprecipitation with the cell extract of undifferentiated PC12 cells. A monoclonal antibody against NDEL1 was used for immunoprecipitation. (E) Amino acids 802–835 of DISC1 are critical for DISC1–NDEL1 interaction. HEK293 cells were transfected with myc-tagged NDEL1 and HA-tagged DISC1 or DISC1 lacking amino acids 802–835 [DISC1Δ(802–835)]. Lysates were then immunoprecipitated with an anti-myc antibody and analyzed by western blotting with an anti-HA antibody. Deletion of amino acids 802–835 of DISC1, which is a critical domain for DISC1–NDEL1 interaction we previously reported, completely abolished the interaction of exogenous DISC1 [DISC1Δ(802–835)] with NDEL1 (top panel). The inputs of each protein are shown (bottom panels). (F) Amino acids 802–835 of DISC1 are critical for DISC1–NDE1 interaction. Myc-tagged NDE1 was transfected with HA-tagged DISC1 or HA-tagged DISC1Δ(802–835) in HEK293 cells. Protein extracts were immunoprecipitated with an anti-myc antibody. Deletion of amino acids 802–835 of DISC1 dramatically weakened the interaction with NDEL1 in co-immunoprecipitation (top panel). The inputs are shown (bottom panels). (G) Protein expression of NDE1 and NDEL1 in the cerebral cortex. NDE1 protein was expressed constantly at a low level from E14 to adulthood. Expression of NDEL1 is increased according to brain development, with the highest expression at P7.
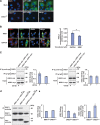
Figure 6.
Possible competition of NDE1 and NDEL1 for DISC1. (A) Change in subcellular distribution of NDE1 by overexpression of DISC1. Exogenous expression of DISC1 (DISC1-Ser704) (red) leads to punctuated distribution of GFP-tagged NDE1 (green) in COS7 cells. Scale bar, 10 µm. (B) NDE1 affects the accumulation of NDEL1 to the centrosome in PC12 cells. Endogenous NDEL1 (red) is localized in the perinuclear region, including the centrosome, in undifferentiated PC12 cells. Overexpression of NDE1 (green, upper panels), but not GAPDH (green, lower panels), redistributes endogenous NDEL1 (red) from the perinuclear regions (lower panels). To semi-quantify the localization change, immunointensity of NDEL1 in the centrosome area (white circle) relative to that in the whole cell region surrounded by green line was examined. Bars represent averages of each group of cells in blinded experiments (*P < 0.005). Error bars represent SEM. Representative images are shown. Blue, nucleus; red, NDEL1; green, NDE1 or GAPDH; white, γ-tubulin (indicated by arrowheads). Scale bar, 10 µm. (C) Protein interactions of DISC1–NDE1 and DISC1–NDEL1 were not affected in the presence of U0126, which is known to block NGF-induced activation of ERK1/2 in PC12 cells. Error bars represent SEM. (D) HEK 293 cells were transfected with HA-tagged DISC1Ser or DISC1Cys together with myc-tagged NDE1. Lysates were then immunoprecipitated with an anti-myc antibody and western-blotted with an antibody to HA to analyze the DISC1–NDE1 interaction. Interaction of DISC1Ser with NDE1 is slightly stronger than that with DISC1Cys as shown in co-immunoprecipitation with an anti-myc antibody (top panel). The inputs are also shown (bottom panels). (E) Influence of the Ser704Cys variation of DISC1 on the DISC1–NDEL1 and DISC1–NDE1 protein interactions. NDE1 binds to DISC1Ser more tightly than to DISC1Cys (left graph and D), whereas NDEL1 interacts with DISC1Ser less tightly than with DISC1Cys as published previously (middle graph). The opposite binding pattern for NDE1 and NDEL1 with regard to DISC1 Ser704Cys results in about 1.6-fold difference between DISC1Ser and DISC1Cys (right graph) (P = 0.054). Error bars represent SEM.
Similar articles
- NDE1 and NDEL1: multimerisation, alternate splicing and DISC1 interaction.
Bradshaw NJ, Christie S, Soares DC, Carlyle BC, Porteous DJ, Millar JK. Bradshaw NJ, et al. Neurosci Lett. 2009 Jan 16;449(3):228-33. doi: 10.1016/j.neulet.2008.10.095. Epub 2008 Nov 6. Neurosci Lett. 2009. PMID: 19000741 Free PMC article. - PKA phosphorylation of NDE1 is DISC1/PDE4 dependent and modulates its interaction with LIS1 and NDEL1.
Bradshaw NJ, Soares DC, Carlyle BC, Ogawa F, Davidson-Smith H, Christie S, Mackie S, Thomson PA, Porteous DJ, Millar JK. Bradshaw NJ, et al. J Neurosci. 2011 Jun 15;31(24):9043-54. doi: 10.1523/JNEUROSCI.5410-10.2011. J Neurosci. 2011. PMID: 21677187 Free PMC article. - Evidence of statistical epistasis between DISC1, CIT and NDEL1 impacting risk for schizophrenia: biological validation with functional neuroimaging.
Nicodemus KK, Callicott JH, Higier RG, Luna A, Nixon DC, Lipska BK, Vakkalanka R, Giegling I, Rujescu D, St Clair D, Muglia P, Shugart YY, Weinberger DR. Nicodemus KK, et al. Hum Genet. 2010 Apr;127(4):441-52. doi: 10.1007/s00439-009-0782-y. Hum Genet. 2010. PMID: 20084519 - DISC1-binding proteins in neural development, signalling and schizophrenia.
Bradshaw NJ, Porteous DJ. Bradshaw NJ, et al. Neuropharmacology. 2012 Mar;62(3):1230-41. doi: 10.1016/j.neuropharm.2010.12.027. Epub 2010 Dec 31. Neuropharmacology. 2012. PMID: 21195721 Free PMC article. Review. - NDE1 and NDEL1 from genes to (mal)functions: parallel but distinct roles impacting on neurodevelopmental disorders and psychiatric illness.
Bradshaw NJ, Hayashi MA. Bradshaw NJ, et al. Cell Mol Life Sci. 2017 Apr;74(7):1191-1210. doi: 10.1007/s00018-016-2395-7. Epub 2016 Oct 14. Cell Mol Life Sci. 2017. PMID: 27742926 Free PMC article. Review.
Cited by
- Molecular links between mitochondrial dysfunctions and schizophrenia.
Park C, Park SK. Park C, et al. Mol Cells. 2012 Feb;33(2):105-10. doi: 10.1007/s10059-012-2284-3. Epub 2012 Feb 15. Mol Cells. 2012. PMID: 22358509 Free PMC article. Review. - Disrupted-in-schizophrenia 1 (DISC1) plays essential roles in mitochondria in collaboration with Mitofilin.
Park YU, Jeong J, Lee H, Mun JY, Kim JH, Lee JS, Nguyen MD, Han SS, Suh PG, Park SK. Park YU, et al. Proc Natl Acad Sci U S A. 2010 Oct 12;107(41):17785-90. doi: 10.1073/pnas.1004361107. Epub 2010 Sep 28. Proc Natl Acad Sci U S A. 2010. PMID: 20880836 Free PMC article. - Molecular characterization of disrupted in schizophrenia-1 risk variant S704C reveals the formation of altered oligomeric assembly.
Narayanan S, Arthanari H, Wolfe MS, Wagner G. Narayanan S, et al. J Biol Chem. 2011 Dec 23;286(51):44266-44276. doi: 10.1074/jbc.M111.271593. Epub 2011 Oct 13. J Biol Chem. 2011. PMID: 21998303 Free PMC article. - DISC1 Regulates Neurogenesis via Modulating Kinetochore Attachment of Ndel1/Nde1 during Mitosis.
Ye F, Kang E, Yu C, Qian X, Jacob F, Yu C, Mao M, Poon RYC, Kim J, Song H, Ming GL, Zhang M. Ye F, et al. Neuron. 2017 Dec 6;96(5):1041-1054.e5. doi: 10.1016/j.neuron.2017.10.010. Epub 2017 Nov 2. Neuron. 2017. PMID: 29103808 Free PMC article. - Common functional polymorphisms of DISC1 and cortical maturation in typically developing children and adolescents.
Raznahan A, Lee Y, Long R, Greenstein D, Clasen L, Addington A, Rapoport JL, Giedd JN. Raznahan A, et al. Mol Psychiatry. 2011 Sep;16(9):917-26. doi: 10.1038/mp.2010.72. Epub 2010 Jul 13. Mol Psychiatry. 2011. PMID: 20628343 Free PMC article.
References
- Porteous D.J., Thomson P., Brandon N.J., Millar J.K. The genetics and biology of DISC1—an emerging role in psychosis and cognition. Biol. Psychiatry. 2006;60:123–131. - PubMed
- Ishizuka K., Paek M., Kamiya A., Sawa A. A review of Disrupted-In-Schizophrenia-1 (DISC1): neurodevelopment, cognition, and mental conditions. Biol. Psychiatry. 2006;59:1189–1197. - PubMed
- Hashimoto R., Numakawa T., Ohnishi T., Kumamaru E., Yagasaki Y., Ishimoto T., Mori T., Nemoto K., Adachi N., Izumi A., et al. Impact of the DISC1 Ser704Cys polymorphism on risk for major depression, brain morphology and ERK signaling. Hum. Mol. Genet. 2006;15:3024–3033. - PubMed
- Callicott J., Straub R.E., Pezawas L., Egan M.F., Mattay V.S., Hariri A.R., Verchinski B.A., Meyer-Lindenberg A., Balkissoon R., Kolachana B., et al. Variation in DISC1 affects hippocampal structure and function and increases risk for schizophrenia. Proc. Natl Acad. Sci. USA. 2004;102:8627–8632. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- K23 MH077807/MH/NIMH NIH HHS/United States
- K99 MH086756/MH/NIMH NIH HHS/United States
- R00 MH086756/MH/NIMH NIH HHS/United States
- Z01 AA000301/ImNIH/Intramural NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical