MicroRNAs are tightly associated with RNA-induced gene silencing complexes in vivo - PubMed (original) (raw)
MicroRNAs are tightly associated with RNA-induced gene silencing complexes in vivo
Fuchou Tang et al. Biochem Biophys Res Commun. 2008.
Abstract
Previous work has shown that synthesized siRNA/miRNA is tightly associated with RNA-induced Gene Silencing Complexes (RISCs) in vitro. However, it is unknown if the endogenous miRNAs are also stably bound to RISC complexes in vivo in cells under physiological conditions. Here we describe the use of the looped real-time PCR-based method to trace the location of endogenous miRNAs in intact cells. We found that most of the endogenous miRNAs are tightly bound to RISC complexes, and only a very small proportion of them are free in cells. Furthermore, synthesized single-stranded mature miRNA or hairpin miRNA precursor cannot replace endogenous miRNAs already present in RISC complexes. However, we found that modified 2-O-Methyl-ribonucleotides were able to dissociate the target miRNA specifically from the RISC complex. These findings have important implications for understanding the basis for the stability and metabolism of miRNAs in living cells.
Figures
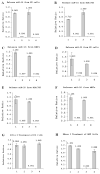
Figure 1. Most miRNA are tightly associated with RISCs.
(A-C) Measurement of miR-16 in ES cells, NIH/3T3 cells, and mouse embryonic fibroblasts (MEFs). (1) Three freeze-thaw cycles followed by 95°C for 5min; (2) Three freeze-thaw cycles only (3) Treatment at 95°C for 5min; (4) Treatment at 4°C only as a control. (D-F) Measurement of miR-20 level in ES cells, NIH/3T3 cells, and MEFs. Treatments 1, 2, 3 and 4 are as specified in A-C. (G) RNase I treatment of ES cell lysates. (1) ES cells after three freeze-thaw cycles, were treated with RNase I for 5min, and following exposure at 95°C for 5min to release all RISC-bond miRNAs. (2) Three freeze-thaw cycles followed by incubation in the buffer for 5min, and at 95°C for 5min to release all RISC-bond miRNAs (as a control for RNase I treatment). (3) Incubation at 95°C for 5min to release miRNAs from RISC complex, followed by RNase I treatment for 5min, and further incubation of the cell lysate at 95°C for 5min. (4) ES cells were incubated at 95°C for 5min to release miRNAs from RISC complex, followed by treatment with buffer treatment for 5min, and incubation at 95°C for 5min (as a control for RNase I treatment). (H) RNase I treatment of MEF (Mouse Embryonic Fibroblast) lysates. (1) MEFs after three freeze-thaw cycles, were treated with RNase I for 5min, and following exposure at 95°C for 5min to release all RISC-bond miRNAs. (2) Three freeze-thaw cycles followed by incubation in the buffer for 5min, and at 95°C for 5min to release all RISC-bond miRNAs (as a control for RNase I treatment). (3) Incubation at 95°C for 5min to release miRNAs from RISC complex, followed by RNase I treatment for 5min, and further incubation of the cell lysate at 95°C for 5min. (4) MEFs were incubated at 95°C for 5min to release miRNAs from RISC complex, followed by treatment with buffer treatment for 5min, and incubation at 95°C for 5min (as a control for RNase I treatment).
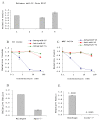
Figure 2. The influence of antagomirs, Ago2, and Dicer on miRNA.
(A) Excessive amounts of synthetic miRNA or its precursor cannot replace endogenous miRNAs-RISCs. (1) ES cells treated at 95°C for 5min to release miRNAs (positive control). (2) Treatment at 95°C for 5min followed by incubation with RNase I for 30min to degrade ‘free’ miRNAs, then 95°C for 5min to release RISC-bond miRNAs (as negative control). (3) ES cells subjected to freeze-thaw cycles, followed by incubation with 0.5uM synthesized let-7a for 30min, and then with RNase I for 30min to degrade ‘free’ miRNAs, and finally 95°C for 5min to release RISC-bond miRNAs. (4) ES cell lysate incubated with 0.5uM synthesized pre-let-7a for 30min, and then with RNase I for 30min to degrade ‘free’ miRNAs, and finally at 95°C for 5min to release RISC-bond miRNAs. (B-C) Antagomirs effect on complementary miRNAs. Cells (ES or MEFs) were lysed by 3 freeze-thaw cycles. Then resulting cell lysate was incubated with 0.5nM-50nM of antagomirs of miR-16 (antagomir-16) for 30min, or as a control, with 0.5nM-50nM antisense miR-16 RNA or an unrelated antagomirs of Let-7a. RNase I was added subsequently to the samples to degrade miRNAs that were released from RISC complexes. Finally, RNase I was inactivated and the remaining RISC-bound miRNAs were released by treatment of the cell lysate at 95°C for 5min. (D) Measurement of mir-16 expression in wildtype and Ago2-/- mouse embryonic fibroblasts (MEFs). (E) Measurement of mir-16 expression in wildtype and Dicer-/- mouse embryonic fibroblasts (MEFs).
Similar articles
- Effective Anti-miRNA Oligonucleotides Show High Releasing Rate of MicroRNA from RNA-Induced Silencing Complex.
Ariyoshi J, Matsuyama Y, Kobori A, Murakami A, Sugiyama H, Yamayoshi A. Ariyoshi J, et al. Nucleic Acid Ther. 2017 Oct;27(5):303-308. doi: 10.1089/nat.2017.0663. Epub 2017 Sep 6. Nucleic Acid Ther. 2017. PMID: 28876213 - Clarifying mammalian RISC assembly in vitro.
Tan GS, Garchow BG, Liu X, Metzler D, Kiriakidou M. Tan GS, et al. BMC Mol Biol. 2011 Apr 29;12:19. doi: 10.1186/1471-2199-12-19. BMC Mol Biol. 2011. PMID: 21529364 Free PMC article. - The Ago2-miRNA-co-IP Assay to Study TGF- β1 Mediated Recruitment of miRNA to the RISC in CFBE Cells.
Mitash N, Donovan JE, Swiatecka-Urban A. Mitash N, et al. J Vis Exp. 2020 Jul 31;(161):10.3791/61571. doi: 10.3791/61571. J Vis Exp. 2020. PMID: 32894261 Free PMC article. - siRNA and miRNA: an insight into RISCs.
Tang G. Tang G. Trends Biochem Sci. 2005 Feb;30(2):106-14. doi: 10.1016/j.tibs.2004.12.007. Trends Biochem Sci. 2005. PMID: 15691656 Review. - Making RISC.
Kawamata T, Tomari Y. Kawamata T, et al. Trends Biochem Sci. 2010 Jul;35(7):368-76. doi: 10.1016/j.tibs.2010.03.009. Epub 2010 Apr 13. Trends Biochem Sci. 2010. PMID: 20395147 Review.
Cited by
- MicroRNAs as Potential Biomarkers in the Differential Diagnosis of Lipomatous Tumors and Their Mimics.
Tan HM, Cheng H, Tang YC, Leong SM, Teo PY, Lee CK, Lee VKM, Hue SS. Tan HM, et al. Int J Mol Sci. 2022 Jul 15;23(14):7804. doi: 10.3390/ijms23147804. Int J Mol Sci. 2022. PMID: 35887151 Free PMC article. - Barley Seeds miRNome Stability during Long-Term Storage and Aging.
Puchta M, Groszyk J, Małecka M, Koter MD, Niedzielski M, Rakoczy-Trojanowska M, Boczkowska M. Puchta M, et al. Int J Mol Sci. 2021 Apr 21;22(9):4315. doi: 10.3390/ijms22094315. Int J Mol Sci. 2021. PMID: 33919202 Free PMC article. - A comprehensive expression profile of micrornas in rat's pituitary.
Yuan B, Han DX, Dai LS, Gao Y, Ding Y, Yu XF, Chen J, Jiang H, Chen CZ, Zhang JB. Yuan B, et al. Int J Clin Exp Med. 2015 Aug 15;8(8):13289-95. eCollection 2015. Int J Clin Exp Med. 2015. PMID: 26550255 Free PMC article. - Reconfigurable hybrid interface for molecular marker diagnostics and in-situ reporting.
Ehrhardt K, Guinn MT, Quarton T, Zhang MQ, Bleris L. Ehrhardt K, et al. Biosens Bioelectron. 2015 Dec 15;74:744-50. doi: 10.1016/j.bios.2015.07.035. Epub 2015 Jul 17. Biosens Bioelectron. 2015. PMID: 26210472 Free PMC article. - MicroRNAs: The Missing Link in the Biology of Graft-Versus-Host Disease?
Atarod S, Dickinson AM. Atarod S, et al. Front Immunol. 2013 Dec 2;4:420. doi: 10.3389/fimmu.2013.00420. Front Immunol. 2013. PMID: 24348483 Free PMC article. Review.
References
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. - PubMed
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. - PubMed
- Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. - PubMed
- Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Bio. 2005;6:376–385. - PubMed
- Pham JW, Pellino JL, Lee YS, Carthew RW, Sontheimer EJ. A Dicer-2-dependent 80s complex cleaves targeted mRNAs during RNAi in Drosophila. Cell. 2004;117:83–94. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources