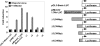The bHLH/Per-Arnt-Sim transcription factor SIM2 regulates muscle transcript myomesin2 via a novel, non-canonical E-box sequence - PubMed (original) (raw)
The bHLH/Per-Arnt-Sim transcription factor SIM2 regulates muscle transcript myomesin2 via a novel, non-canonical E-box sequence
Susan Woods et al. Nucleic Acids Res. 2008 Jun.
Abstract
Despite a growing number of descriptive studies that show Single-minded 2 (Sim2) is not only essential for murine survival, but also upregulated in colon, prostate and pancreatic tumours, there is a lack of direct target genes identified for this basic helix-loop-helix/PAS transcription factor. We have performed a set of microarray experiments aimed at identifying genes that are differentially regulated by SIM2, and successfully verified that the Myomesin2 (Myom2) gene is SIM2-responsive. Although SIM2 has been reported to be a transcription repressor, we find that SIM2 induces transcription of Myom2 and activates the Myom2 promoter sequence when co-expressed with the heterodimeric partner protein, ARNT1, in human embryonic kidney cells. Truncation and mutation of the Myom2 promoter sequence, combined with chromatin immunoprecipitation studies in cells, has lead to the delineation of a non-canonical E-box sequence 5'-AACGTG-3' that is bound by SIM2/ARNT1 heterodimers. Interestingly, in immortalized human myoblasts knock down of Sim2 results in increased levels of Myom2 RNA, suggesting that SIM2 is acting as a repressor in these cells and so its activity is likely to be highly context dependent. This is the first report of a direct SIM2/ARNT1 target gene with accompanying analysis of a functional response element.
Figures
Figure 1.
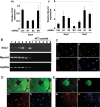
Myom2 promoter is induced by SIM2/AD expression. Delineation of required promoter regions and a potential SIM2 responsive site. Subconfluent 293T cells were co-transfected with luciferase reporter plasmids and a blank or SIM2/AD expression plasmid as indicated. After 48 h luciferase activity was assayed and relative luciferase activities normalized to the value for each reporter alone and presented as a fold induction. Results depicted are from an experiment performed in triplicate with standard deviation, which is representative of four repeats of this set of transfections.
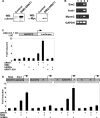
Figure 2.
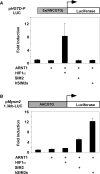
Expression of the SIM2/ARNT1 heterodimer induces Myom2 transcription in human embryonic kidney cells through a non-canonical E-box. (A) Nuclear extracts prepared from the 293Control or 293SIM2/ARNT1 cell lines were subjected to SDS–PAGE and western blot using either anti-ARNT1 polyclonal or anti-Myc monoclonal antibodies. (B) total RNA samples prepared from untransfected 293a cells (lane 1), 293Control (lane 2) or 293SIM2/ARNT1 (lane 3) double stable cells, were reverse transcribed and the resultant cDNA samples were PCR-amplified in the linear range using primers specific for Sim2, Arnt1, Myom2 and the internal control, GAPDH. Control Myom2 reactions containing RNA samples as template were also performed (lane 4). C and D, 293 cells were co-transfected for 48h with the _Myom2_-promoter containing reporter plasmid and expression vectors as indicated and then assayed for luciferase activity. Luciferase activity was normalized against a Renilla luciferase internal control and results are depicted as fold induction over the backbone pGL3-Basic-LUC reporter for each transfection. Results shown are either from a single experiment performed in triplicate with standard deviation, which is representative of four repeats of this set of transfections (C) or from three independent experiments performed in triplicate with standard deviation (D).
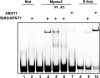
Figure 3.
SIM2/ARNT1 dimers recognise the 40 bp Myom2 promoter sequence in vitro. Bacterially co-expressed and partially purified, thioredoxin/6xHis tagged N-terminal portions of SIM2 and ARNT1 or ARNT1 alone were bound to fluorescently labelled 40bp Myom2, Myom2mut or E-box probes and subjected to non-denaturing PAGE. The presence of ARNT1 in the gel shifted complex was visualized by incubation with anti-ARNT1 serum (A1) compared to control pre-immune serum (PI).
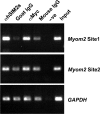
Figure 4.
The endogenous Myom2 promoter is bound by SIM2 in human cells. Chromatin extracts from 293 cells stably expressing ARNT1 and Myc-epitope tagged hSIM2s were immunoprecipitated with antibodies recognising hSIM2s (αhSIM2s) or the Myc tag (αMyc) or an equal amount of the non-specific immunoglobulin controls (Goat IgG and Mouse IgG). DNA eluted from the chromatin immunoprecipitations was examined by PCR amplification using primers designed to amplify the Site 1 or Site 3 (containing an identical hexamer site to Site 1 but 850 bp upstream) sequences from the Myom2 promoter or the control GAPDH sequence. Input contains DNA isolated from the chromatin extract and –ve indicates a control eluate in which no chromatin extract was added to the immunoprecipitation.
Figure 5.
SIM2 represses Myom2 expression in human myoblasts but is coexpressed with MYOM2 in adult mouse muscle. (A) Knockdown of Sim2 in LHCN-M2 myoblasts results in increased Myom2 expression. Level of Sim2 (i) and Myom2 (ii) RNA in LHCN-M2 cells treated with control (ctrl), Myom2 siRNA (M1) or either of two independent Sim2 siRNAs (S1 and S2) determined by real-time PCR. Experiment is representative of a set of three repeats, Sim2 and Myom2 expression is relative to GAPDH expression and error bars depict standard deviation. (B) Total RNA samples prepared from adult mouse muscle tissues (lane 1 abdominal muscle, lane 2 diaphragm, lane 3 forelimb muscle, lane 4 hindlimb muscle, lane 5 heart) and kidney (lane 6) were reverse transcribed and the resultant cDNA samples were PCR-amplified in the linear range using primers specific for Sim2, Myom2 and the internal control, GAPDH. (C–E) Multiple muscle fibres are presented in the figure in cross section and each fibre is surrounded by multiple nuclei. (C) SIM2 is expressed in myonuclei of adult mouse muscle. Composite image (i) of cryostat sections of adult mouse hind limb muscle immunostained for myogenin (ii) and SIM2 proteins (iii), showing nuclei visualised with bis-Benzimide H (iv). (D) SIM2 is expressed in both MYOM2 positive and negative muscle fibres. Composite image (i) of cryostat sections of adult mouse hind limb muscle immunostained for MYOM2 (ii, fast fibres) and SIM2 proteins (iii), showing nuclei visualized with bis-Benzimide H (iv). (E) composite image (i) of cryostat sections of adult mouse hind limb muscle immunostained for EH-MYOM1 (ii, one positive fibre in view) and SIM2 proteins (iii), showing nuclei visualised with bis-Benzimide H (iv).
Figure 6.
The Myom2 promoter is not activated by HIF-1α/ARNT1. 293 cells were cotransfected for 40 h with the p_HGTD_-P_-LUC (A) or p_Myom2_1.3kbBasic-LUC (B) reporter plasmids and expression vectors as indicated and lysates were assayed for luciferase activity. Luciferase activity was normalised against a Renilla luciferase internal control and results are depicted as fold induction over the reporter alone for each transfection. Results depicted are from a single experiment performed in triplicate with standard deviation, which is representative of repeats of this set of transfections.
Similar articles
- Characterization of functionally deficient SIM2 variants found in patients with neurological phenotypes.
Button EL, Rossi JJ, McDougal DP, Bruning JB, Peet DJ, Bersten DC, Rosenfeld JA, Whitelaw ML. Button EL, et al. Biochem J. 2022 Jul 15;479(13):1441-1454. doi: 10.1042/BCJ20220209. Biochem J. 2022. PMID: 35730699 Free PMC article. - Two murine homologs of the Drosophila single-minded protein that interact with the mouse aryl hydrocarbon receptor nuclear translocator protein.
Probst MR, Fan CM, Tessier-Lavigne M, Hankinson O. Probst MR, et al. J Biol Chem. 1997 Feb 14;272(7):4451-7. doi: 10.1074/jbc.272.7.4451. J Biol Chem. 1997. PMID: 9020169 - The mammalian basic helix-loop-helix/PAS family of transcriptional regulators.
Kewley RJ, Whitelaw ML, Chapman-Smith A. Kewley RJ, et al. Int J Biochem Cell Biol. 2004 Feb;36(2):189-204. doi: 10.1016/s1357-2725(03)00211-5. Int J Biochem Cell Biol. 2004. PMID: 14643885 Review. - The aryl hydrocarbon receptor nuclear translocator (ARNT) family of proteins: transcriptional modifiers with multi-functional protein interfaces.
Labrecque MP, Prefontaine GG, Beischlag TV. Labrecque MP, et al. Curr Mol Med. 2013 Aug;13(7):1047-65. doi: 10.2174/15665240113139990042. Curr Mol Med. 2013. PMID: 23116263 Review.
Cited by
- Regulated transcription of human matrix metalloproteinase 13 (MMP13) and interleukin-1β (IL1B) genes in chondrocytes depends on methylation of specific proximal promoter CpG sites.
Hashimoto K, Otero M, Imagawa K, de Andrés MC, Coico JM, Roach HI, Oreffo ROC, Marcu KB, Goldring MB. Hashimoto K, et al. J Biol Chem. 2013 Apr 5;288(14):10061-10072. doi: 10.1074/jbc.M112.421156. Epub 2013 Feb 15. J Biol Chem. 2013. PMID: 23417678 Free PMC article. - Identification of the transcription factor single-minded homologue 2 as a potential biomarker and immunotherapy target in prostate cancer.
Arredouani MS, Lu B, Bhasin M, Eljanne M, Yue W, Mosquera JM, Bubley GJ, Li V, Rubin MA, Libermann TA, Sanda MG. Arredouani MS, et al. Clin Cancer Res. 2009 Sep 15;15(18):5794-802. doi: 10.1158/1078-0432.CCR-09-0911. Epub 2009 Sep 8. Clin Cancer Res. 2009. PMID: 19737960 Free PMC article. - Characterization of functionally deficient SIM2 variants found in patients with neurological phenotypes.
Button EL, Rossi JJ, McDougal DP, Bruning JB, Peet DJ, Bersten DC, Rosenfeld JA, Whitelaw ML. Button EL, et al. Biochem J. 2022 Jul 15;479(13):1441-1454. doi: 10.1042/BCJ20220209. Biochem J. 2022. PMID: 35730699 Free PMC article. - Overexpression of PGC-1α in aging muscle enhances a subset of young-like molecular patterns.
Garcia S, Nissanka N, Mareco EA, Rossi S, Peralta S, Diaz F, Rotundo RL, Carvalho RF, Moraes CT. Garcia S, et al. Aging Cell. 2018 Apr;17(2):e12707. doi: 10.1111/acel.12707. Epub 2018 Feb 10. Aging Cell. 2018. PMID: 29427317 Free PMC article. - Role of developmental factors in hypothalamic function.
Biran J, Tahor M, Wircer E, Levkowitz G. Biran J, et al. Front Neuroanat. 2015 Apr 21;9:47. doi: 10.3389/fnana.2015.00047. eCollection 2015. Front Neuroanat. 2015. PMID: 25954163 Free PMC article. Review.
References
- Crews ST, Fan CM. Remembrance of things PAS: regulation of development by bHLH-PAS proteins. Curr. Opin. Genet. Dev. 1999;9:580–587. - PubMed
- Panda S, Hogenesch JB, Kay SA. Circadian rhythms from flies to human. Nature. 2002;417:329–335. - PubMed
- Kewley RJ, Whitelaw ML, Chapman-Smith A. The mammalian basic helix-loop-helix/PAS family of transcriptional regulators. Int. J. Biochem. Cell. Biol. 2004;36:189–204. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases