Gli3 coordinates three-dimensional patterning and growth of the tectum and cerebellum by integrating Shh and Fgf8 signaling - PubMed (original) (raw)
. 2008 Jun;135(12):2093-103.
doi: 10.1242/dev.015990. Epub 2008 May 14.
Affiliations
- PMID: 18480159
- PMCID: PMC2673693
- DOI: 10.1242/dev.015990
Gli3 coordinates three-dimensional patterning and growth of the tectum and cerebellum by integrating Shh and Fgf8 signaling
Sandra Blaess et al. Development. 2008 Jun.
Abstract
The coordination of anterior-posterior (AP) and dorsal-ventral (DV) patterning of the mesencephalon (mes) and rhombomere 1 (r1) is instrumental for the development of three distinct brain structures: the tectum and cerebellum dorsally and the tegmentum ventrally. Patterning of the mes/r1 is primarily mediated by signaling molecules secreted from two organizers: sonic hedgehog (Shh) from the floor plate (DV) and Fgf8 from the isthmus (AP). Gli3, a zinc-finger transcription factor in the Shh signaling pathway, has been implicated in regulating Fgf8 expression and is therefore a potential candidate for coordinating the action of the two organizers. By inactivating mouse Gli3 at successive embryonic time points in vivo, we uncovered the extent and the underlying mechanism of Gli3 function in the mes/r1. We demonstrate that before E9.0, Gli3 is required for establishing a distinct posterior tectum, isthmus and cerebellum, but does not play a role in the development of the tegmentum. Between E9.0 and E11.0, Gli3 continues to be required for isthmus and cerebellum development, but primarily for defining the cerebellar foliation pattern. We show that Gli3 regulates patterning of the isthmus and cerebellar anlage by confining Fgf8 expression to the isthmus, and attenuates growth of dorsal r1 (before E11.0) and the dorsal mes and isthmus (beyond E11.0) through regulation of cell proliferation and viability. In conclusion, our results show that Gli3 is essential for the coordinated three-dimensional patterning and growth of the dorsal mes/r1.
Figures
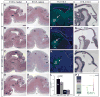
Fig. 1. Distinct temporal roles of Gli3 in regulating midbrain and cerebellum development
(A–H) Hematoxylin and Eosin (H+E) staining of midline (A–D) and lateral (E–H) E18.5 sagittal brain sections. (B,F) In Gli3_−/_− mutants, the dorsal midbrain is enlarged and the distinct morphology of the inferior (Ic) and superior colliculus (Sc) is lost. Similarly, the isthmus (Is) and cerebellum (Cb) are not clearly separated and contain cell clusters (red arrowheads). The Cb is not foliated. The morphology of the ventral mid/hindbrain (vMh) appears normal. (C,G) In En1-Gli3 cko mutants, the Sc, Ic, Is and Cb (arrow) are enlarged, and tectum, Is and Cb are morphological distinct from one another. The Cb foliation pattern is abnormal. (D,H) In Nes-Gli3 cko mutants, the Sc, Ic, Is and Cb are morphologically distinct, but the Sc, Ic and Is are increased in size. (I–K) Immunohistochemistry for tyrosine hydroxylase (TH) shows no change in dopaminergic neurons (green, arrows) in the mutants. DAPI staining is in blue. (L) Quantitative assessment of Cb and Sc size in WT and Nes-Gli3 cko brains as means of samples from three different animals +/− SEM. Student’s _t_-test was performed. (M–O) H+E staining of E10.5 sagittal embryo sections. Note the increased size of the ventricle and increased thickness and abnormal morphology of the Is/r1 region in Gli3_−/_− mutants and En1-Gli3 cko mutants. (P) Left: Schematic of Shh and Gli expression in the ventral (V) and dorsal (D) embryonic mes/r1. Right: Shh signaling in the ventral and dorsal mes/r1: High levels of Shh induce Gli activator (GliA2/3; green) and inhibit (red) the formation of Gli3 repressor ventrally (Gli3R, purple). Low levels of Shh decrease the formation of Gli3R dorsally. Gradients indicate high to low levels of expression/signaling. Scale bar: (A–H) 500 μm; (I–K, M–N) 250 μm
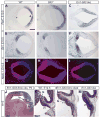
Fig. 2. Gli3 is required for proper establishment of the inferior colliculus
(A–D) En1 RNA expression in the E18.5 inferior colliculus (Ic) and posterior superior colliculus (Sc) in WT sagittal sections (A). En1 expression is severely reduced in Gli3_−/_− mutants, slightly reduced in En1-Gli3 cko and normal in Nes-Gli3 cko mutants (B–D). (E–H) Expression of Otx2 RNA in superficial layers of the Sc and the ventricular layer of the Ic is comparable in WT (E) and Nes-Gli3 cko brains (H). In Gli3_−/_− and En1-Gli3 cko mutants Otx2 is expressed throughout the posterior tectum and the thickness of the Otx2 positive layer is increased in the Sc (F,G). (I,J) Immunohistochemistry for Neurogranin on E18.5 sagittal sections. The Neurogranin positive domain (outlined in red) is reduced and shifted posteriorly in Gli3_−/_− mutants. (K–N) DAPI staining (K) and immunohistochemistry for Neurogranin (L–N) at P16 shows that the Ic is abnormally shaped in En1-Gli3 cko, but not in Nes-Gli3 cko mutants. Scale bars: (A–H,I,J,L–N) 250 μm; (K) 500 μm.
Fig. 3. Gli3 regulates proper establishment of the isthmus and cerebellum
(A–E) RNA in situ hybridization for Math1 and DAPI staining shows that the external granule cell layer (EGL) is absent form the most posterior and anterior parts (arrowheads) of the isthmus-cerebellar like (Is/Cb) region in Gli3_−/_− mutants, but is comparable to WT in En1-Gli3 cko mutants (arrowheads). (F–N) Immunohistochemistry on adjacent sections. (F–K) In the WT, Calbindin and RORα (green) positive Purkinje cells (PC) are organized in a several-cell-deep layer (PCL) underlying the EGL and project into the deeper Cb (arrowheads). In Gli3_−/_− mutants, only a rudimentary PCL forms with disorganized projections (arrowheads), and many PC remain in clusters in the deeper Is/Cb (outlined). (H,K) In En1-Gli3 cko mutants, most PC are located within the PCL, with only some scattered PC in the underlying areas (H′,K′, arrowheads) and in ectopic clusters in the anterior Is (outlined). Some PC axons project into the Is (H, arrowhead). (L–M) Pax2 (green) is expressed in a scattered pattern throughout the Is and Cb (except the EGL and PCL) in WT and En1-Gli3 cko mutants but is not expressed in the anterior (EGL-free) region in Gli3_−/_− mutants and is excluded from the PC clusters (M, arrowheads). (O–Q) H+E staining of P2 sagittal sections shows the abnormal foliation pattern in En1-Gli3 cko mutants. Brain regions are outlined where necessary. Note that some of the presented pictures are composites of two images (C,D,F,G,I,J) Scale bars: (A–N) 200 μm; (H′, K′) 20 μm; (P–Q) 500 μm
Fig. 4. Gli3 is not required to establish DV gene expression domains or to inhibit activating Shh signaling
(A–I) RNA in situ hybridization for Pax7 (A–C, dorsal marker) and Gli1 (D–F, ventral marker) and immunohistochemistry for Nkx6.1 on E10.5 transverse sections (G–I, ventral marker) shows that expression of these genes is comparable to WT in Gli3_−/_− and En1-Gli3 cko mutants. (J–M) H+E staining on P0 (J) and E12.5 (K–M) sagittal sections. The phenotype of the Sc, Ic and Is in P0 En1-Gli3;Smo cko mutants is comparable to En1-Gli3 cko mutants. Note that the Cb is small and unfoliated, with a thin external granule cell layer. (K–M) At E12.5, the size of r1 is increased in both En1-Gli3 and En1-Gli3;Smo cko mutants compared to WT. The neural tube is outlined where necessary. v (ventral), d (dorsal). Scale bars: (A–I) 125 μm, (J) 500 μm, (K–M) 200 μm.
Fig. 5. Gli3 is not required to establish the mes/r1 roof plate
(A–F) RNA in situ hybridization for Gdf7 and Wnt1 on E9.5 transverse sections. Gdf7 and Wnt1 are expressed in the roof plate (RP) in the WT and mutant embryos. Note that the Wnt1 positive domain in the lateral mes (*) is in the isthmic region. (G–J) RNA in situ hybridization for Msx1 and Axin2 on E10.5 transverse sections shows that RP expression is not changed in Gli3_−/_− mutants. (K) Plane of sections are indicated in the schematic. The neural tube is outlined where necessary. Scale bars: (A–J) 100 μm.
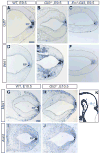
Fig. 6. Gli3 is required to restrict Fgf expression to the isthmus
Fgf8, Fgf17, Spry1 and Wnt1 RNA expression. Posterior mes, Is and r1 are shown (see Fig. 5K). Black arrowheads indicate normal, red arrowheads ectopic gene expression. (A–F) The Wnt1 expression domain is unaltered in Gli3 mutants. Fgf8 (G–I), Fgf17 (M–O), and Spry1 (SU) domains are expanded into medial, but not lateral r1 in E10.5 Gli3_−/− mutants. In E12.5 En1-Gli3 cko mutants, ectopic expression of Fgf8 (J,K), Fgf17_ (P,Q), and Spry1 (V,W) is restricted to the most posterior region of medial r1, where Wnt1 (D,E) is normally expressed. (L,R,X) Fgf8, Fgf17, and Spry1 gene expression is normal in Nes-Gli3 cko mutants. Scale bars: 200 μm.
Fig. 7. Partial rescue of Gli3_−/_− mutant phenotype in Gli3_−/−; Fgf8+/_− mutants
(A) H+E staining on Gli3_−/−;Fgf8+/_− mutant sagittal sections. The morphology of the Cb and Is, but not of the tectum (Sc and Ic), appears to be partially rescued in Gli3_−/−;Fgf8+/_− mutants. (C–D) En1 and Otx2 RNA expression and immunohistochemistry for Neurogranin (D, red outline) show that the Ic is not properly established in Gli3_−/−;Fgf8+/_− mutants. (E,F) Math1 RNA expression (E) and DAPI staining (F, blue) show that the EGL expands from the posterior Cb to the Is (arrowheads), comparable to WT. (F,G) Immunohistochemistry for Calbindin and RORα (green) show a relative normal PCL, but a significant number of PCs are located in clusters in the deeper Cb and Is (G, outlined). (H) Pax2 (green) positive cells are found in the Is, but are excluded from PC clusters (arrowhead). Scale bars: 200 μm.
Fig. 8. The distinct temporal roles of Gli3R in regulating mes/r1 development
(A) Time period of Gli3 gene expression, prenatal tectum and cerebellum phenotype and ectopic Fgf8 expression. Note that in Gli3_−/_− mutants, a domain (X) forms between the tectum and cerebellum that is not properly specified as cerebellum (Cb), isthmus (Is) or inferior colliculus (Ic). Superior colliculus (Sc). See discussion for details (B) High levels of Shh (lower pathway) regulate mes/r1 growth through induction of proliferation through Gli2A and/or inhibition of cell death through Gli3R. Low levels of Shh (upper pathway) do not induce proliferation, but modulate cell death and proliferation (induced by unknown signal (X)) through the regulation of Gli3R levels.
Similar articles
- The duration of Fgf8 isthmic organizer expression is key to patterning different tectal-isthmo-cerebellum structures.
Sato T, Joyner AL. Sato T, et al. Development. 2009 Nov;136(21):3617-26. doi: 10.1242/dev.041210. Epub 2009 Sep 30. Development. 2009. PMID: 19793884 Free PMC article. - Sonic hedgehog regulates Gli activator and repressor functions with spatial and temporal precision in the mid/hindbrain region.
Blaess S, Corrales JD, Joyner AL. Blaess S, et al. Development. 2006 May;133(9):1799-809. doi: 10.1242/dev.02339. Epub 2006 Mar 29. Development. 2006. PMID: 16571630 - Suppressor of fused controls mid-hindbrain patterning and cerebellar morphogenesis via GLI3 repressor.
Kim JJ, Gill PS, Rotin L, van Eede M, Henkelman RM, Hui CC, Rosenblum ND. Kim JJ, et al. J Neurosci. 2011 Feb 2;31(5):1825-36. doi: 10.1523/JNEUROSCI.2166-10.2011. J Neurosci. 2011. PMID: 21289193 Free PMC article. - Essential roles of Gli3 and sonic hedgehog in pattern formation and developmental anomalies caused by their dysfunction.
Motoyama J. Motoyama J. Congenit Anom (Kyoto). 2006 Sep;46(3):123-8. doi: 10.1111/j.1741-4520.2006.00114.x. Congenit Anom (Kyoto). 2006. PMID: 16922918 Review. - Isthmus organizer for midbrain and hindbrain development.
Nakamura H, Katahira T, Matsunaga E, Sato T. Nakamura H, et al. Brain Res Brain Res Rev. 2005 Sep;49(2):120-6. doi: 10.1016/j.brainresrev.2004.10.005. Epub 2005 Jan 21. Brain Res Brain Res Rev. 2005. PMID: 16111543 Review.
Cited by
- Permanent deconstruction of intracellular primary cilia in differentiating granule cell neurons.
Ott CM, Constable S, Nguyen TM, White K, Lee WA, Lippincott-Schwartz J, Mukhopadhyay S. Ott CM, et al. J Cell Biol. 2024 Oct 7;223(10):e202404038. doi: 10.1083/jcb.202404038. Epub 2024 Aug 13. J Cell Biol. 2024. PMID: 39137043 Free PMC article. - SIRV: spatial inference of RNA velocity at the single-cell resolution.
Abdelaal T, Grossouw LM, Pasterkamp RJ, Lelieveldt BPF, Reinders MJT, Mahfouz A. Abdelaal T, et al. NAR Genom Bioinform. 2024 Aug 6;6(3):lqae100. doi: 10.1093/nargab/lqae100. eCollection 2024 Sep. NAR Genom Bioinform. 2024. PMID: 39108639 Free PMC article. - Dual and opposing roles for the kinesin-2 motor, KIF17, in Hedgehog-dependent cerebellar development.
Waas B, Carpenter BS, Franks NE, Merchant OQ, Verhey KJ, Allen BL. Waas B, et al. Sci Adv. 2024 Apr 26;10(17):eade1650. doi: 10.1126/sciadv.ade1650. Epub 2024 Apr 26. Sci Adv. 2024. PMID: 38669326 Free PMC article. - Intervertebral disc-intrinsic Hedgehog signaling maintains disc cell phenotypes and prevents disc degeneration through both cell autonomous and non-autonomous mechanisms.
Zhang L, Hu S, Xiu C, Li M, Zheng Y, Zhang R, Li B, Chen J. Zhang L, et al. Cell Mol Life Sci. 2024 Feb 3;81(1):74. doi: 10.1007/s00018-023-05106-x. Cell Mol Life Sci. 2024. PMID: 38308696 Free PMC article. - Modulation of canonical Wnt signaling regulates peribiliary mesenchymal identity during homeostasis and injury.
Singh S, Budiman T, Redmond D, Gupta V. Singh S, et al. Hepatol Commun. 2024 Jan 22;8(2):e0368. doi: 10.1097/HC9.0000000000000368. eCollection 2024 Feb 1. Hepatol Commun. 2024. PMID: 38251878 Free PMC article.
References
- Agarwala S, Sanders TA, Ragsdale CW. Sonic hedgehog control of size and shape in midbrain pattern formation. Science. 2001;291:2147–50. - PubMed
- Alder J, Lee KJ, Jessell TM, Hatten ME. Generation of cerebellar granule neurons in vivo by transplantation of BMP-treated neural progenitor cells. Nat Neurosci. 1999;2:535–40. - PubMed
- Alvarez-Medina R, Cayuso J, Okubo T, Takada S, Marti E. Wnt canonical pathway restricts graded Shh/Gli patterning activity through the regulation of Gli3 expression. Development. 2008;135:237–47. - PubMed
- Aoto K, Nishimura T, Eto K, Motoyama J. Mouse GLI3 regulates Fgf8 expression and apoptosis in the developing neural tube, face, and limb bud. Dev Biol. 2002;251:320–32. - PubMed
- Bai CB, Auerbach W, Lee JS, Stephen D, Joyner AL. Gli2, but not Gli1, is required for initial Shh signaling and ectopic activation of the Shh pathway. Development. 2002;129:4753–61. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA128158/CA/NCI NIH HHS/United States
- R01 CA128158-12/CA/NCI NIH HHS/United States
- R01 HD035768/HD/NICHD NIH HHS/United States
- R01 HD035768-10/HD/NICHD NIH HHS/United States
- R01 HD035768-09/HD/NICHD NIH HHS/United States
- R01 HD050767-05/HD/NICHD NIH HHS/United States
- R01 HD050767/HD/NICHD NIH HHS/United States
- R01 CA128158-11/CA/NCI NIH HHS/United States
- R01 HD050767-04/HD/NICHD NIH HHS/United States
- R01 HD035768-11/HD/NICHD NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous