HapMap tagSNP transferability in multiple populations: general guidelines - PubMed (original) (raw)
HapMap tagSNP transferability in multiple populations: general guidelines
Jinchuan Xing et al. Genomics. 2008 Jul.
Abstract
Linkage disequilibrium (LD) has received much attention recently because of its value in localizing disease-causing genes. Due to the extensive LD between neighboring loci in the human genome, it is believed that a subset of the single nucleotide polymorphisms in a region (tagSNPs) can be selected to capture most of the remaining SNP variants. In this study, we examined LD patterns and HapMap tagSNP transferability in more than 300 individuals. A South Indian sample and an African Mbuti Pygmy population sample were included to evaluate the performance of HapMap tagSNPs in geographically distinct and genetically isolated populations. Our results show that HapMap tagSNPs selected with r(2) >= 0.8 can capture more than 85% of the SNPs in populations that are from the same continental group. Combined tagSNPs from HapMap CEU and CHB+JPT serve as the best reference for the Indian sample. The HapMap YRI are a sufficient reference for tagSNP selection in the Pygmy sample. In addition to our findings, we reviewed over 25 recent studies of tagSNP transferability and propose a general guideline for selecting tagSNPs from HapMap populations.
Figures
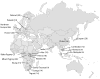
Figure 1. Populations examined
Number of individuals in each population sample is given in parentheses.
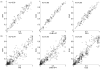
Figure 2. Correlations of allele frequencies (A) and LD measures (_r_2) for all SNP pairs (B) between HapMap populations and corresponding continental groups
Spearman’s correlations (rho) are shown.
Figure 3. Correlation of allele frequency between HapMap populations and A) Indians; B) Mbuti Pygmies
Spearman’s correlations (rho) are shown.
Figure 4. Correlation of pairwise LD (_r_2) between HapMap populations and A) Indians; B) Mbuti Pygmies
Spearman’s correlations (rho) are shown.
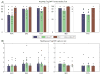
Figure 5. HapMap tagSNP transferability and tagging efficiency
(A) HapMap tagSNP transferability in three continental groups (AFR, EAS and EUR) and two populations (IND and PYG) are shown. The average transferability among all 14 regions are shown as bars, and the transferability for each individual region are shown as black dots. For example, the first blue bar in the “AFR” section indicates that tagSNPs selected from the HapMap CEU population captured ~60% of the SNPs with _r_2 >= 0.8 in our Africans, on average. (B) HapMap tagSNP tagging efficiency. The average tagging efficiency across all 14 regions are shown as bars, and the tagging efficiencies for each region are shown as black dots. For example, the last brown bar in the “PYG” section indicates that on average every HapMap YRI tagSNPs captured 1.21 SNPs in our African Pygmy samples.
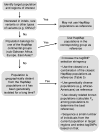
Figure 6
A flow chart for tagSNP selection using HapMap populations.
Similar articles
- TagSNP transferability and relative loss of variability prediction from HapMap to an admixed population.
Lins TC, Abreu BS, Pereira RW. Lins TC, et al. J Biomed Sci. 2009 Aug 14;16(1):73. doi: 10.1186/1423-0127-16-73. J Biomed Sci. 2009. PMID: 19682379 Free PMC article. - On transferability of genome-wide tagSNPs.
Gu CC, Yu K, Ketkar S, Templeton AR, Rao DC. Gu CC, et al. Genet Epidemiol. 2008 Feb;32(2):89-97. doi: 10.1002/gepi.20269. Genet Epidemiol. 2008. PMID: 17896344 Review. - Linkage disequilibrium patterns and tagSNP transferability among European populations.
Mueller JC, Lõhmussaar E, Mägi R, Remm M, Bettecken T, Lichtner P, Biskup S, Illig T, Pfeufer A, Luedemann J, Schreiber S, Pramstaller P, Pichler I, Romeo G, Gaddi A, Testa A, Wichmann HE, Metspalu A, Meitinger T. Mueller JC, et al. Am J Hum Genet. 2005 Mar;76(3):387-98. doi: 10.1086/427925. Epub 2005 Jan 6. Am J Hum Genet. 2005. PMID: 15637659 Free PMC article. - The portability of tagSNPs across populations: a worldwide survey.
González-Neira A, Ke X, Lao O, Calafell F, Navarro A, Comas D, Cann H, Bumpstead S, Ghori J, Hunt S, Deloukas P, Dunham I, Cardon LR, Bertranpetit J. González-Neira A, et al. Genome Res. 2006 Mar;16(3):323-30. doi: 10.1101/gr.4138406. Epub 2006 Feb 8. Genome Res. 2006. PMID: 16467560 Free PMC article. - Evaluating HapMap SNP data transferability in a large-scale genotyping project involving 175 cancer-associated genes.
Ribas G, González-Neira A, Salas A, Milne RL, Vega A, Carracedo B, González E, Barroso E, Fernández LP, Yankilevich P, Robledo M, Carracedo A, Benítez J. Ribas G, et al. Hum Genet. 2006 Feb;118(6):669-79. doi: 10.1007/s00439-005-0094-9. Epub 2005 Dec 2. Hum Genet. 2006. PMID: 16323010 Review.
Cited by
- Identifying highly conserved and highly differentiated gene ontology categories in human populations.
Jiang Y, Zhang R, Sun P, Tang G, Zhang X, Wang X, Guo X, Wang Q, Li X. Jiang Y, et al. PLoS One. 2011;6(11):e27871. doi: 10.1371/journal.pone.0027871. Epub 2011 Nov 30. PLoS One. 2011. PMID: 22140477 Free PMC article. - Genotype-imputation accuracy across worldwide human populations.
Huang L, Li Y, Singleton AB, Hardy JA, Abecasis G, Rosenberg NA, Scheet P. Huang L, et al. Am J Hum Genet. 2009 Feb;84(2):235-50. doi: 10.1016/j.ajhg.2009.01.013. Am J Hum Genet. 2009. PMID: 19215730 Free PMC article. - Genetic diversity in India and the inference of Eurasian population expansion.
Xing J, Watkins WS, Hu Y, Huff CD, Sabo A, Muzny DM, Bamshad MJ, Gibbs RA, Jorde LB, Yu F. Xing J, et al. Genome Biol. 2010;11(11):R113. doi: 10.1186/gb-2010-11-11-r113. Epub 2010 Nov 24. Genome Biol. 2010. PMID: 21106085 Free PMC article. - Human genetic variation and its contribution to complex traits.
Frazer KA, Murray SS, Schork NJ, Topol EJ. Frazer KA, et al. Nat Rev Genet. 2009 Apr;10(4):241-51. doi: 10.1038/nrg2554. Nat Rev Genet. 2009. PMID: 19293820 Review. - Shades of gray: a comparison of linkage disequilibrium between Hutterites and Europeans.
Thompson EE, Sun Y, Nicolae D, Ober C. Thompson EE, et al. Genet Epidemiol. 2010 Feb;34(2):133-9. doi: 10.1002/gepi.20442. Genet Epidemiol. 2010. PMID: 19697328 Free PMC article.
References
- Hästbacka J, de la Chapelle A, Mahtani MM, Clines G, Reeve-Daly MP, Daly M, Hamilton BA, et al. The diastrophic dysplasia gene encodes a novel sulfate transporter: positional cloning by fine-structure linkage disequilibrium mapping. Cell. 1994;78:1073–1087. - PubMed
- Puffenberger EG, Kauffman ER, Bolk S, Matise TC, Washington SS, Angrist M, Weissenbach J, et al. Identity-by-descent and association mapping of a recessive gene for Hirschsprung disease on human chromosome 13q22. Hum Molec Genet. 1994;8:1217–1225. - PubMed
- Feder JN, Gnirke A, Thomas W, Tsuchihashi Z, Ruddy DA, Basava A, Dormishian F, Domingo R, Jr, Ellis MC, Fullan A, Hinton LM, Jones NL, Kimmel BE, Kronmal GS, Lauer P, Lee VK, Loeb DB, Mapa FA, McClelland E, Meyer NC, Mintier GA, Moeller N, Moore T, Morikang E, Wolff RK, et al. A novel MHC class I-like gene is mutated in patients with hereditary haemochromatosis. Nat Genet. 1996;13:399–408. - PubMed
- Jorde LB. Linkage disequilibrium and the search for complex disease genes. Genome Res. 2000;10:1435–44. - PubMed
Publication types
MeSH terms
Grants and funding
- R01 GM059290-07/GM/NIGMS NIH HHS/United States
- HL-070048/HL/NHLBI NIH HHS/United States
- R01 GM059290/GM/NIGMS NIH HHS/United States
- GM-59290/GM/NIGMS NIH HHS/United States
- R01 HL070048/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Research Materials