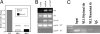MLL protects CpG clusters from methylation within the Hoxa9 gene, maintaining transcript expression - PubMed (original) (raw)
MLL protects CpG clusters from methylation within the Hoxa9 gene, maintaining transcript expression
Frank E Erfurth et al. Proc Natl Acad Sci U S A. 2008.
Abstract
Homeobox (HOX) genes play a definitive role in determination of cell fate during embryogenesis and hematopoiesis. MLL-related leukemia is coincident with increased expression of a subset of HOX genes, including HOXA9. MLL functions to maintain, rather than initiate, expression of its target genes. However, the mechanism of MLL maintenance of target gene expression is not understood. Here, we demonstrate that Mll binds to specific clusters of CpG residues within the Hoxa9 locus and regulates expression of multiple transcripts. The presence of Mll at these clusters provides protection from DNA methylation. shRNA knock-down of Mll reverses the methylation protection status at the previously protected CpG clusters; methylation at these CpG residues is similar to that observed in Mll null cells. Furthermore, reconstituting MLL expression in Mll null cells can reverse DNA methylation of the same CpG residues, demonstrating a dominant effect of MLL in protecting this specific region from DNA methylation. Intriguingly, an oncogenic MLL-AF4 fusion can also reverse DNA methylation, but only for a subset of these CpGs. This method of transcriptional regulation suggests a mechanism that explains the role of Mll in transcriptional maintenance, but it may extend to other CpG DNA binding proteins. Protection from methylation may be an important mechanism of epigenetic inheritance by regulating the function of both de novo and maintenance DNA methyltransferases.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
MLL protects specific CpG residues in the Hoxa9 locus from methylation. (A) Schematic representation of the murine Hoxa9 region. Eight kilobases of the genomic region of Hoxa9 is represented. CpG Islands are numbered beneath. Upstream alternative first exon is labeled AB, the canonical Hoxa9 first exon is labeled CD and the canonical second exon, containing the homeodomain, is labeled II. Large asterisks indicate Mll binding as determined by ChIP. (B) MLL-dependent protection of CpG Island 1 from methylation. Direct bisulfite sequencing reveals a difference in methylation status in Mll+/+ MEF cells (circles) and _Mll_−/− MEF cells (X). Area under the curve analysis was performed on DNA sequence histograms and relative methylation percentage was determined for CpG residues. Each point on the graph represents a single CpG residue from the sequence below the graph. Beneath the graph are sequencing results from individual clones (10 clones each from Mll+/+ and _Mll_−/− MEFs) of PCR products from bisulfite treated template. Empty circles, unmethylated CpG residues; filled circles, methylated CpG residues. (C) Upon shRNA knockdown of Mll in Mll+/+ MEF cells, protection from methylation is lost. Triangles, shRNA Mll knockdown; circles, shRNA pSuper vector alone. Knockdowns were selected for 2 weeks. Four-week selection showed similar results (data not shown). (D) Upon add-back of MLL in _Mll_−/− MEF cells, protection from methylation returns. Filled circles, MLL add-back; x's, vector-only control. Data shown are from cells kept under selection for one week.
Fig. 2.
Mll-dependent expression of Hoxa9 transcripts and Mll binding to Hoxa9 AB region. (A) RNA from Mll+/+ and _Mll_−/− MEF cells was analyzed by real-time RT-PCR to detect expression of upstream AB and canonical Hoxa9 transcripts. Expression was normalized to gapdh from the same samples. (B) shRNA knockdown of Mll. RNA extracted from Mll+/+ MEF cells after infection with retrovirus carrying empty vector or shRNA directed against either Mll or CtBP (an unrelated gene) were analyzed by RT-PCR to detect expression of Mll (to confirm efficient knockdown), the upstream AB transcript, the canonical Hoxa9 transcript, or gapdh control with and without reverse transcription (± RT). Cells were kept under selection for 2 weeks. (C) ChIP analysis was performed by using a mixture of polyclonal anti-Mll antibodies, a commercial anti-Mll monoclonal antibody, or IgG control antibody. Enriched chromatin was amplified by using primers specific for the AB exon.
Fig. 3.
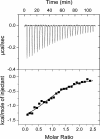
ITC measurement of the binding of the MLL CXXC domain to CpG1II oligonucleotide. Data shown is for addition of 10-μl aliquots of 550 μM CXXC domain to a 46 μM solution of CpG1II. The data were fit to a one-site binding model, giving _K_d = 7.8 ± 1.4 μM and n = 0.93 ± 0.04.
Fig. 4.
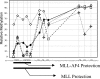
Add back of MLL-AF4 fusion reduces methylation of a portion of the protected CpG1 residues after 4 weeks and longer. _Mll_−/− MEF cells were transfected with either plasmid to express MLL-AF4 or empty vector. Cell populations were analyzed after 4 weeks of selection (squares, MLL-AF4; diamonds, vector control) or individual clones were selected, replated, and expanded (crosses and circles, two different representative MLL-AF4 add-back clones). DNA was subjected to bisulfite treatment and nested PCR and sequenced. Area under the curve analysis was performed on DNA sequencing histograms and relative methylation percentage was determined for CpG residues. Regions protected from DNA methylation by MLL-AF4 and by MLL (data from Fig. 1) are highlighted under the graph.
Similar articles
- Functional specificity of CpG DNA-binding CXXC domains in mixed lineage leukemia.
Risner LE, Kuntimaddi A, Lokken AA, Achille NJ, Birch NW, Schoenfelt K, Bushweller JH, Zeleznik-Le NJ. Risner LE, et al. J Biol Chem. 2013 Oct 11;288(41):29901-10. doi: 10.1074/jbc.M113.474858. Epub 2013 Aug 29. J Biol Chem. 2013. PMID: 23990460 Free PMC article. - Down-regulation of DLX3 expression in MLL-AF4 childhood lymphoblastic leukemias is mediated by promoter region hypermethylation.
Campo Dell'Orto M, Banelli B, Giarin E, Accordi B, Trentin L, Romani M, te Kronnie G, Basso G. Campo Dell'Orto M, et al. Oncol Rep. 2007 Aug;18(2):417-23. Oncol Rep. 2007. PMID: 17611665 - MLL targets SET domain methyltransferase activity to Hox gene promoters.
Milne TA, Briggs SD, Brock HW, Martin ME, Gibbs D, Allis CD, Hess JL. Milne TA, et al. Mol Cell. 2002 Nov;10(5):1107-17. doi: 10.1016/s1097-2765(02)00741-4. Mol Cell. 2002. PMID: 12453418 - Leukemogenesis via aberrant self-renewal by the MLL/AEP-mediated transcriptional activation system.
Yokoyama A. Yokoyama A. Cancer Sci. 2021 Oct;112(10):3935-3944. doi: 10.1111/cas.15054. Epub 2021 Aug 2. Cancer Sci. 2021. PMID: 34251718 Free PMC article. Review. - Molecular mechanisms of MLL-associated leukemia.
Yokoyama A. Yokoyama A. Int J Hematol. 2015 Apr;101(4):352-61. doi: 10.1007/s12185-015-1774-4. Epub 2015 Mar 17. Int J Hematol. 2015. PMID: 25773519 Review.
Cited by
- Hematopoietic Differentiation of Human Pluripotent Stem Cells: HOX and GATA Transcription Factors as Master Regulators.
Alsayegh K, Cortés-Medina LV, Ramos-Mandujano G, Badraiq H, Li M. Alsayegh K, et al. Curr Genomics. 2019 Sep;20(6):438-452. doi: 10.2174/1389202920666191017163837. Curr Genomics. 2019. PMID: 32194342 Free PMC article. Review. - Modes of Interaction of KMT2 Histone H3 Lysine 4 Methyltransferase/COMPASS Complexes with Chromatin.
Bochyńska A, Lüscher-Firzlaff J, Lüscher B. Bochyńska A, et al. Cells. 2018 Mar 2;7(3):17. doi: 10.3390/cells7030017. Cells. 2018. PMID: 29498679 Free PMC article. Review. - DNA Methylation in the Exon 1 Region and Complex Regulation of Twist1 Expression in Gastric Cancer Cells.
Sakamoto A, Akiyama Y, Shimada S, Zhu WG, Yuasa Y, Tanaka S. Sakamoto A, et al. PLoS One. 2015 Dec 22;10(12):e0145630. doi: 10.1371/journal.pone.0145630. eCollection 2015. PLoS One. 2015. PMID: 26695186 Free PMC article. - MicroRNAs in leukemias: emerging diagnostic tools and therapeutic targets.
Mian YA, Zeleznik-Le NJ. Mian YA, et al. Curr Drug Targets. 2010 Jul;11(7):801-11. doi: 10.2174/138945010791320872. Curr Drug Targets. 2010. PMID: 20370647 Free PMC article. Review. - RNAi-mediated silencing of MLL-AF9 reveals leukemia-associated downstream targets and processes.
Fleischmann KK, Pagel P, Schmid I, Roscher AA. Fleischmann KK, et al. Mol Cancer. 2014 Feb 11;13:27. doi: 10.1186/1476-4598-13-27. Mol Cancer. 2014. PMID: 24517546 Free PMC article.
References
- Popovic R, Zeleznik-Le NJ. MLL: How complex does it get? J Cell Biochem. 2005;95:234–242. - PubMed
- Armstrong SA, et al. MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat Genet. 2002;30:41–47. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials