Cell-surface protein-protein interaction analysis with time-resolved FRET and snap-tag technologies: application to GPCR oligomerization - PubMed (original) (raw)
doi: 10.1038/nmeth.1213. Epub 2008 May 18.
Laëtitia Comps-Agrar, Carsten Brock, Marie-Laure Rives, Emmanuel Bourrier, Mohammed Akli Ayoub, Hervé Bazin, Norbert Tinel, Thierry Durroux, Laurent Prézeau, Eric Trinquet, Jean-Philippe Pin
Affiliations
- PMID: 18488035
- PMCID: PMC2642604
- DOI: 10.1038/nmeth.1213
Cell-surface protein-protein interaction analysis with time-resolved FRET and snap-tag technologies: application to GPCR oligomerization
Damien Maurel et al. Nat Methods. 2008 Jun.
Abstract
Cell-surface proteins are important in cell-cell communication. They assemble into heterocomplexes that include different receptors and effectors. Elucidation and manipulation of such protein complexes offers new therapeutic possibilities. We describe a methodology combining time-resolved fluorescence resonance energy transfer (FRET) with snap-tag technology to quantitatively analyze protein-protein interactions at the surface of living cells, in a high throughput-compatible format. Using this approach, we examined whether G protein-coupled receptors (GPCRs) are monomers or assemble into dimers or larger oligomers--a matter of intense debate. We obtained evidence for the oligomeric state of both class A and class C GPCRs. We also observed different quaternary structure of GPCRs for the neurotransmitters glutamate and gamma-aminobutyric acid (GABA): whereas metabotropic glutamate receptors assembled into strict dimers, the GABA(B) receptors spontaneously formed dimers of heterodimers, offering a way to modulate G-protein coupling efficacy. This approach will be useful in systematic analysis of cell-surface protein interaction in living cells.
Figures
Figure 1
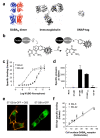
The use of snap-tag to label surface proteins with TR-FRET compatible fluorophores. (a) Ribbon representations at the same scale of the heterodimeric GABAB receptor (left, as based on the structure of the dimer of mGlu1 extracellular domains (pdb accession number: 1EWK), and the structure of a possible rhodopsin dimer (pdb: 1N3M)), an IgG (center, pdb: 1IGT) and a snap-tag (right, pdb 1EH6). (b) Covalent labeling of snap-tag fusion protein using 0-benzyl guanine derivatives carrying a fluorophore (F). (c) Cell surface specific binding with increasing concentrations of BG-K (filled circles) or BG-d2 (open circles) on ST-GABAB1 co-expressed with GABAB2. (d) Specific BG-K labeling of mock transfected cells, and cells expressing the ST-GABAB1 alone (ST-GB1) or co-expressed with GABAB2 (ST-GB1:GB2), ST-GABAB1 mutated in its intracellular retention motif (ST-GB1ASA), and the G-protein αi1 subunit fused to snap-tag (Gi1-ST). (e) Confocal imaging of cells over-expressing a ST-GABAB1-GFP fusion alone (right) or together with GABAB2 (left) and labeled with BG-d2. (f) Amount of d2 (open circles) and K (closed circles) fluorophores specifically bound to cells expressing various amounts of ST-GABAB receptors at the cell surface. Cells were transfected with a fixed amount of ST-GABAB1 and increasing amounts of GAB AB2 plasmids. The specific number of cell surface receptors (Bmax) was determined by Scatchard analysis using [3H]-CGP54626, a non permeant GABAB1 specific antagonist. Linear regression revealed a slope (number of fluorophores per GABAB dimers) of 1.05 and 1.04 for the BG-K and BG-d2 labeling, respectively. Data in c, d and f are means ± sem of triplicate determinations from a representative experiment.
Figure 2
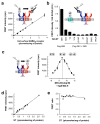
Detection of GABAB heteromers at the cell surface using snap-tag and TR-FRET. (a) FRET intensity between d2-labeled anti-flag antibodies and BG-K labeled snap-tags in cells expressing increasing amounts of surface ST-GABAB1 and flag-GABAB2 receptors. FRET is measured as the specific d2 emission at 665nm after excitation of europium cryptate at 337 nm, the background signal measured in the absence of d2-antibodies being substracted. The FRET intensity is represented according to the total number of receptors expressed at the cell surface, (b) TR-FRET was measured between flag-GABAB receptors labeled with d2-antibodies, and the indicated HA-snap-tag fusion proteins labeled with BG-K. Data were obtained with the same amount of snap-tag proteins at the cell surface as measured with anti-anti-HA ELISA, and a constant amount of flag-GABAB receptors. Positive control (left column) was performed with cells expressing ST-GABAB1 and flag-GABAB2. (c) FRET intensity was measured in cells expressing ST-GABAB1 and ST-GABAB2 with varying concentration of BG-d2 and 5_μ_M BG-K. (d) FRET intensity is directly proportional to the amount of ST-GABA subunits at the cell surface. The number of snap-tags was deduced from the Bmax of [3H]-CGP54626. (e) FRET efficacy as determined by the ratio of the specific d2 emission at 665 nm resulting from FRET, and the fluorescence intensity (at 620 nm) of the specifically bound BG-K, is plotted as a function of the amount of snap-tags at the cell surface deduced from the Bmax of [3H]-CGP54626. Data in (a) and (b) are means ± sem of triplicate determinations from a typical experiments. Data in (d) and (e) are triplicate determinations from 4 independent experiments.
Figure 3
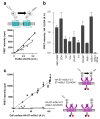
Detection of cell surface protein oligomers using snap-tag fusions and TR-FRET. (a) Both TR-FRET intensity and HA-ELISA were measured for various expression levels of either GABAB (filled squares) or V2 vasopressin (open squares) HA-ST-fusions. (b) Experiments were conducted as in (a) for various other cell surface proteins, and means TR-FRET intensity over the ELISA signal (representing the slope) are presented. Negative control (right column “mGlu1 dimer”) was performed using a mGlu1 receptor dimer carrying a single ST (see (c)). (c) FRET intensity was plotted as a function of the amount of mGlu1 receptor at the cell surface, when both subunits are fused to snap-tag (filled symbols) or when only one subunit per dimer is labeled (open symbols). To control the subunit composition of the mGlu1 receptor dimer, each subunit carries the C-terminal tail of either GABAB1 (C1 in blue) with its natural intracellular retention signal (blue ball) or GABAB2 (C2 in red) in which an intracellular retention signal was added (red ball). Coiled-coil interaction between Cl and C2 prevents intracellular retention of both proteins such that neither subunit reach the surface alone, but do so when co-expressed together (supp Fig. D).
Figure 4
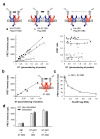
Detection of GABAB dimers of dimers at the cell surface, (a) FRET intensity (left panel) and FRET efficacy (right panel) measured in cells expressing various amounts of the GABAB receptor combinations illustrated in the top schemes: when both subunits carry a snap-tag (filled squares), when only GABAB1 carries a snap-tag (grey circles), or when only GABAB2 carries a snap-tag (open triangles). FRET intensity is plotted as a function of the amount of snap-tags at the cell surface deduced from the Bmax of [3H]-CGP54626. (b) FRET intensity as a function of the amount of ST-GAB AB2 expressed alone, (c) FRET between ST-GABAB2 subunits as a function of the amount of transfected GABAB1 plasmid. (d) GABA activation of the GABAB heteromer affects neither the FRET between GABAB2 subunits (GB1+ST-GB2), nor the FRET between GABAB1 subunits (ST-GB1+GB2), nor the FRET between GABAB1 and GABAB2 subunits (ST-GB1+ST-GB2).
Figure 5
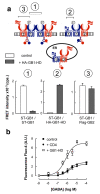
Functional correlate of the association between GABAB heterodimers (a) top: Scheme representing the various combinations of subunits co-expressed: ST-GABAB1(blue) with flag-GAB AB2KKXX (red, the KKXX motif being the intracellular retention sequence added after the coiled coil domain), with (right) or without (left) the HA-GB1-HD, corresponding to the GB1 subunit deleted of both its extracellular and intracellular domains. Bottom: TR-FRET intensities measured between two ST-GABAB1 labeled with BG-K and BG-d2 (left (1)), between ST-GABAB1 labeled with BG-K and HA-GB1-HD labeled with d2-conjugated anti-HA antibodies (middle (2)), and between ST-GABAB1 labeled with BG-K and flag-GAB AB2KKXX labeled with d2-conjugated anti-flag antibodies (right (3)), in the absence (white columns) and in the presence (grey columns) of HA-GB1-HD. b) Ca2+ signals generated in cells expressing ST-GABAB1 and flag-GABAB2KKXX together (filled squares), or in the presence of over-expressed HA-GB1-HD (open squares), or in the presence of over-expressed CD4 (filled circles). Data are means ± sem of triplicate determinations from a typical experiment.
Similar articles
- Cell-surface protein-protein interaction analysis with time-resolved FRET and snap-tag technologies: application to G protein-coupled receptor oligomerization.
Comps-Agrar L, Maurel D, Rondard P, Pin JP, Trinquet E, Prézeau L. Comps-Agrar L, et al. Methods Mol Biol. 2011;756:201-14. doi: 10.1007/978-1-61779-160-4_10. Methods Mol Biol. 2011. PMID: 21870227 - Single-molecule FRET imaging of GPCR dimers in living cells.
Asher WB, Geggier P, Holsey MD, Gilmore GT, Pati AK, Meszaros J, Terry DS, Mathiasen S, Kaliszewski MJ, McCauley MD, Govindaraju A, Zhou Z, Harikumar KG, Jaqaman K, Miller LJ, Smith AW, Blanchard SC, Javitch JA. Asher WB, et al. Nat Methods. 2021 Apr;18(4):397-405. doi: 10.1038/s41592-021-01081-y. Epub 2021 Mar 8. Nat Methods. 2021. PMID: 33686301 Free PMC article. - Cell-surface protein-protein interaction analysis with time-resolved FRET and snap-tag technologies.
Feinstein TN. Feinstein TN. Methods Mol Biol. 2013;1066:121-9. doi: 10.1007/978-1-62703-604-7_11. Methods Mol Biol. 2013. PMID: 23955739 - G-protein-coupled receptor oligomers: two or more for what? Lessons from mGlu and GABAB receptors.
Pin JP, Comps-Agrar L, Maurel D, Monnier C, Rives ML, Trinquet E, Kniazeff J, Rondard P, Prézeau L. Pin JP, et al. J Physiol. 2009 Nov 15;587(Pt 22):5337-44. doi: 10.1113/jphysiol.2009.179978. Epub 2009 Sep 1. J Physiol. 2009. PMID: 19723778 Free PMC article. Review. - Methods used to study the oligomeric structure of G-protein-coupled receptors.
Guo H, An S, Ward R, Yang Y, Liu Y, Guo XX, Hao Q, Xu TR. Guo H, et al. Biosci Rep. 2017 Apr 20;37(2):BSR20160547. doi: 10.1042/BSR20160547. Print 2017 Apr 30. Biosci Rep. 2017. PMID: 28062602 Free PMC article. Review.
Cited by
- Spatially resolved subcellular protein-protein interactomics in drug-perturbed lung-cancer cultures and tissues.
Cai S, Hu T, Venkataraman A, Rivera Moctezuma FG, Ozturk E, Zhang N, Wang M, Zvidzai T, Das S, Pillai A, Schneider F, Ramalingam SS, Oh YT, Sun SY, Coskun AF. Cai S, et al. Nat Biomed Eng. 2024 Oct 30. doi: 10.1038/s41551-024-01271-x. Online ahead of print. Nat Biomed Eng. 2024. PMID: 39478233 - mGlu4R, mGlu7R, and mGlu8R allosteric modulation for treating acute and chronic neurodegenerative disorders.
Domin H, Burnat G. Domin H, et al. Pharmacol Rep. 2024 Sep 30. doi: 10.1007/s43440-024-00657-7. Online ahead of print. Pharmacol Rep. 2024. PMID: 39348087 Review. - Impact of aging on the GABAB receptor-mediated connectome.
Vásquez E, Oresti GM, Paez MD, Callegari EA, Masone D, Muñoz EM. Vásquez E, et al. bioRxiv [Preprint]. 2024 Aug 2:2024.07.31.606013. doi: 10.1101/2024.07.31.606013. bioRxiv. 2024. PMID: 39131332 Free PMC article. Preprint. - Mechanistic insights from the atomic-level quaternary structure of short-lived GPCR oligomers in live cells.
Stoneman MR, Yokoi K, Biener G, Killeen TD, Adhikari DP, Rahman S, Harikumar KG, Miller LJ, Raicu V. Stoneman MR, et al. Res Sq [Preprint]. 2024 Jul 19:rs.3.rs-4683780. doi: 10.21203/rs.3.rs-4683780/v1. Res Sq. 2024. PMID: 39070646 Free PMC article. Preprint. - Bidirectional linkage of DNA barcodes for the multiplexed mapping of higher-order protein interactions in cells.
Liu Y, Sundah NR, Ho NRY, Shen WX, Xu Y, Natalia A, Yu Z, Seet JE, Chan CW, Loh TP, Lim BY, Shao H. Liu Y, et al. Nat Biomed Eng. 2024 Jul;8(7):909-923. doi: 10.1038/s41551-024-01225-3. Epub 2024 Jun 19. Nat Biomed Eng. 2024. PMID: 38898172
References
- Bayburt TH, Leitz AJ, Xie G, Oprian DD, Sligar SG. Transducin activation by nanoscale lipid bilayers containing one and two rhodopsins. J Biol Chem. 2007;282:14875–14881. - PubMed
- Chabre M, le Maire M. Monomeric G-protein-coupled receptor as a functional unit. Biochemistry. 2005;44:9395–9403. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources